1G3M
 
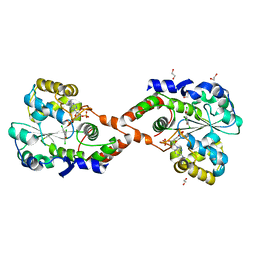 | | CRYSTAL STRUCTURE OF HUMAN ESTROGEN SULFOTRANSFERASE IN COMPLEX WITH IN-ACTIVE COFACTOR PAP AND 3,5,3',5'-TETRACHLORO-BIPHENYL-4,4'-DIOL | | Descriptor: | 1,2-ETHANEDIOL, 3,5,3',5'-TETRACHLORO-BIPHENYL-4,4'-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Shevtsov, S, Petrochenko, E.V, Negishi, M, Pedersen, L.C. | | Deposit date: | 2000-10-24 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic analysis of a hydroxylated polychlorinated biphenyl (OH-PCB) bound to the catalytic estrogen binding site of human estrogen sulfotransferase.
Environ.Health Perspect., 111, 2003
|
|
2C1J
 
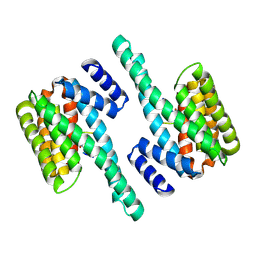 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
1HMO
 
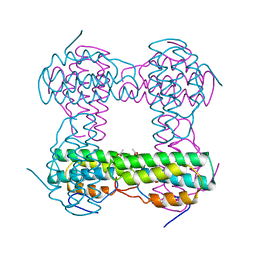 | | THE STRUCTURE OF DEOXY AND OXY HEMERYTHRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ACETYL GROUP, HEMERYTHRIN, MU-OXO-DIIRON, ... | | Authors: | Holmes, M.A, Letrong, I, Turley, S, Sieker, L.C, Stenkamp, R.E. | | Deposit date: | 1990-10-18 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of deoxy and oxy hemerythrin at 2.0 A resolution.
J.Mol.Biol., 218, 1991
|
|
1FDN
 
 | | REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDURICI AT 1.84 ANGSTROMS RESOLUTION | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Duee, E, Fanchon, E, Vicat, J, Sieker, L.C, Meyer, J, Moulis, J-M. | | Deposit date: | 1994-03-31 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution.
J.Mol.Biol., 243, 1994
|
|
1FCN
 
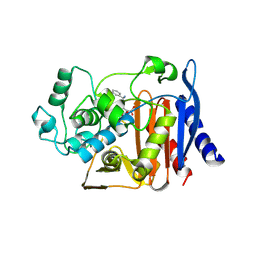 | |
2BCV
 
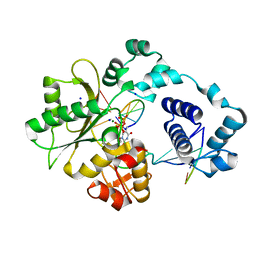 | | DNA polymerase lambda in complex with Dttp and a DNA duplex containing an unpaired Dtmp | | Descriptor: | 5'-D(*CP*AP*GP*TP*AP*(O2C))-3', 5'-D(*CP*GP*GP*CP*AP*GP*TP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Cell(Cambridge,Mass.), 124, 2006
|
|
2BLS
 
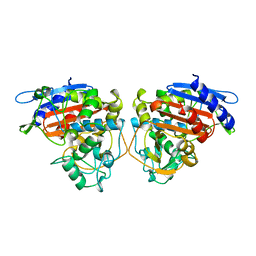 | |
2C1A
 
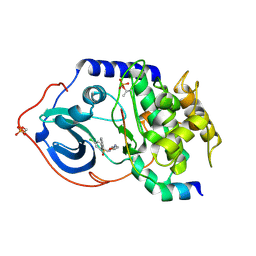 | | Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
1YRK
 
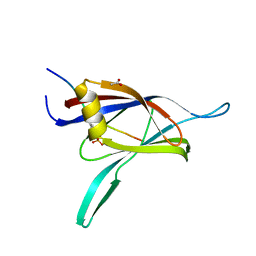 | | The C2 Domain of PKC is a new Phospho-Tyrosine Binding Domain | | Descriptor: | 13-residue peptide, ACETIC ACID, Protein kinase C, ... | | Authors: | Benes, C.H, Wu, N, Elia, A.E, Dharia, T, Cantley, L.C, Soltoff, S.P. | | Deposit date: | 2005-02-03 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C2 domain of PKCdelta is a phosphotyrosine binding domain.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YW2
 
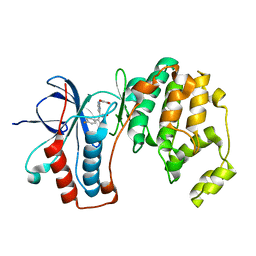 | | Mutated Mus Musculus P38 Kinase (mP38) | | Descriptor: | 2-(ETHOXYMETHYL)-4-(4-FLUOROPHENYL)-3-[2-(2-HYDROXYPHENOXY)PYRIMIDIN-4-YL]ISOXAZOL-5(2H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Laughlin, S.K, Clark, M.P, Djung, J.F, Golebiowski, A, Brugel, T.A, Sabat, M, Bookland, R.G, Laufersweiler, M.J, Vanrens, J.C, Townes, J.A, De, B, Hsieh, L.C, Xu, S.C, Walter, R.L, Mekel, M.J, Janusz, M.J. | | Deposit date: | 2005-02-16 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The development of new isoxazolone based inhibitors of tumor necrosis factor-alpha (TNF-alpha) production.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1IRN
 
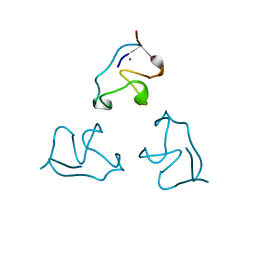 | | RUBREDOXIN (ZN-SUBSTITUTED) AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M, Meyer, J. | | Deposit date: | 1995-12-13 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1ZJN
 
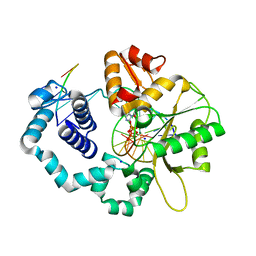 | | Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2005-04-29 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate.
STRUCTURE, 13, 2005
|
|
1G21
 
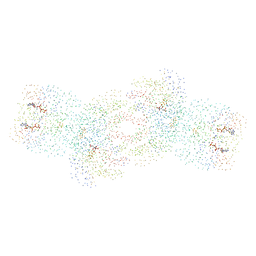 | | MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Chiu, H.-J, Peters, J.W, Lanzilotta, W.N, Ryle, M.J, Seefeldt, L.C, Howard, J.B, Rees, D.C. | | Deposit date: | 2000-10-16 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Biochemistry, 40, 2001
|
|
2C1B
 
 | | Structure of cAMP-dependent protein kinase complexed with (4R,2S)-5'-(4-(4-Chlorobenzyloxy)pyrrolidin-2-ylmethanesulfonyl)isoquinoline | | Descriptor: | (4R,2S)-5'-(4-(4-CHLOROBENZYLOXY)PYRROLIDIN-2-YLMETHANESULFONYL)ISOQUINOLINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
2BFI
 
 | | Molecular basis for amyloid fibril formation and stability | | Descriptor: | SYNTHETIC PEPTIDE | | Authors: | Makin, O.S, Atkins, E, Sikorski, P, Johansson, J, Serpell, L.C. | | Deposit date: | 2004-12-07 | | Release date: | 2005-01-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Basis for Amyloid Fibril Formation and Stability
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1FX1
 
 | |
1ZM8
 
 | | Apo Crystal structure of Nuclease A from Anabaena sp. | | Descriptor: | MANGANESE (II) ION, Nuclease, SODIUM ION, ... | | Authors: | Ghosh, M, Meiss, G, Pingoud, A, London, R.E, Pedersen, L.C. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Mechanism of Nuclease A, a beta beta alpha Metal Nuclease from Anabaena.
J.Biol.Chem., 280, 2005
|
|
1GGY
 
 | | HUMAN FACTOR XIII WITH YTTERBIUM BOUND IN THE ION SITE | | Descriptor: | PROTEIN (COAGULATION FACTOR XIII), YTTERBIUM (III) ION | | Authors: | Fox, B.A, Yee, V.C, Pederson, L.C, Trong, I.L, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1998-07-23 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
1YVC
 
 | | Solution structure of the conserved protein from the gene locus MMP0076 of Methanococcus maripaludis. Northeast Structural Genomics target MrR5. | | Descriptor: | MrR5 | | Authors: | Northeast Structural Genomics Consortium (NESG), Rossi, P, Aramini, J.M, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T. | | Deposit date: | 2005-02-15 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: |
To be Published
|
|
1YWM
 
 | | Crystal structure of the N-terminal domain of group B Streptococcus alpha C protein | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C protein alpha-antigen, GLYCEROL | | Authors: | Auperin, T.C, Bolduc, G.R, Baron, M.J, Heroux, A, Filman, D.J, Madoff, L.C, Hogle, J.M. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the N-terminal domain of the group B streptococcus alpha C protein.
J.Biol.Chem., 280, 2005
|
|
2C0X
 
 | | MOLECULAR STRUCTURE OF FD FILAMENTOUS BACTERIOPHAGE REFINED WITH RESPECT TO X-RAY FIBRE DIFFRACTION AND SOLID-STATE NMR DATA | | Descriptor: | COAT PROTEIN B | | Authors: | Marvin, D.A, Welsh, L.C, Symmons, M.F, Scott, W.R.P, Straus, S.K. | | Deposit date: | 2005-09-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-Ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane.
J.Mol.Biol., 355, 2006
|
|
1GGT
 
 | | THREE-DIMENSIONAL STRUCTURE OF A TRANSGLUTAMINASE: HUMAN BLOOD COAGULATION FACTOR XIII | | Descriptor: | COAGULATION FACTOR XIII | | Authors: | Yee, V.C, Pedersen, L.C, Trong, I.L, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1994-01-25 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
2BX5
 
 | | Is FR1 the antibody's Achillies heel | | Descriptor: | VD9 VKI LIGHT-CHAIN | | Authors: | James, L.C. | | Deposit date: | 2005-07-22 | | Release date: | 2006-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beta-Edge Interactions in a Pentadecameric Human Antibody Vkappa Domain.
J.Mol.Biol., 367, 2007
|
|
1ZGI
 
 | | thrombin in complex with an oxazolopyridine inhibitor 21 | | Descriptor: | (R)-2-(2-(1H-1,2,4-TRIAZOL-1-YL)BENZYL)-N-(2,2-DIFLUORO-2-(PIPERIDIN-2-YL)ETHYL)OXAZOLO[4,5-C]PYRIDIN-4-AMINE, Hirudin, Thrombin | | Authors: | Deng, J.Z, McMasters, D.R, Rabbat, P.M, Williams, P.D, Coburn, C.A, Yan, Y, Kuo, L.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Strulovici, B, Vacca, J.P, Lyle, T.A, Burgey, C.S. | | Deposit date: | 2005-04-21 | | Release date: | 2005-09-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of an oxazolopyridine series of dual thrombin/factor Xa inhibitors via structure-guided lead optimization.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1IFQ
 
 | | Sec22b N-terminal domain | | Descriptor: | GLYCEROL, vesicle trafficking protein Sec22b | | Authors: | Gonzalez Jr, L.C, Weis, W.I, Scheller, R.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel snare N-terminal domain revealed by the crystal structure of Sec22b.
J.Biol.Chem., 276, 2001
|
|
