4USC
 
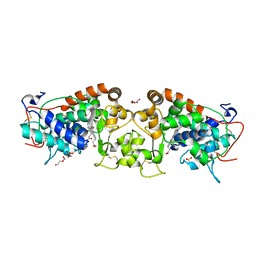 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
6OKJ
 
 | |
1KEM
 
 | |
5IGN
 
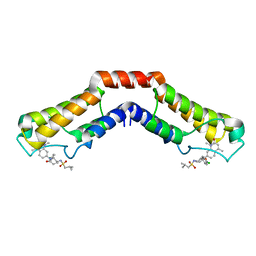 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
4V1P
 
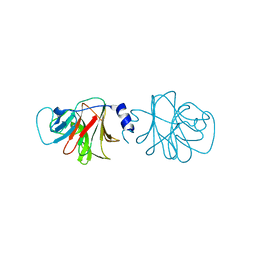 | | BTN3 Structure | | Descriptor: | BUTYROPHILIN SUBFAMILY 3 MEMBER A1 | | Authors: | James, L.C. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Activation of Human Gammadelta T Cells by Cytosolic Interactions of Btn3A1 with Soluble Phosphoantigens and the Cytoskeletal Adaptor Periplakin.
J.Immunol., 194, 2015
|
|
7DLX
 
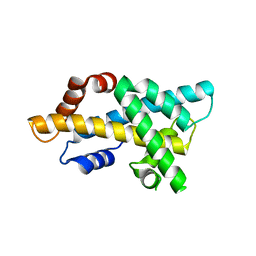 | | crystal structure of H2AM4>Z-H2B | | Descriptor: | Histone H2B,Histone H2A | | Authors: | Dai, L.C, Zhou, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Recognition of the inherently unstable H2A nucleosome by Swc2 is a major determinant for unidirectional H2A.Z exchange.
Cell Rep, 35, 2021
|
|
6XUV
 
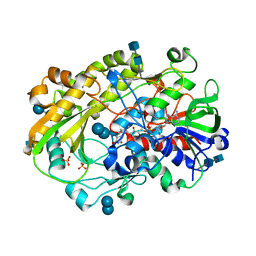 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, laminaribiose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6XUU
 
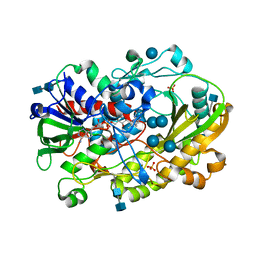 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, glucose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
8OWA
 
 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-4 | | Descriptor: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | Authors: | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
8OWL
 
 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-6 | | Descriptor: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | Authors: | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
2P0Y
 
 | | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR6 | | Descriptor: | Hypothetical protein lp_0780 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Chi, K.H, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum.
To be Published
|
|
2P7N
 
 | | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69. | | Descriptor: | Pathogenicity island 1 effector protein | | Authors: | Benach, J, Abashidze, M, Seetharaman, J, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69.
To be Published
|
|
2DO8
 
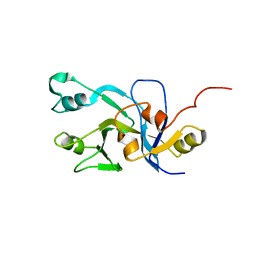 | | Solution Structure of UPF0301 protein HD_1794 | | Descriptor: | UPF0301 protein HD_1794 | | Authors: | Zhang, Q, Liu, G, Shastry, R, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T.R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of UPF0301 protein HD_1794
To be published
|
|
6ES8
 
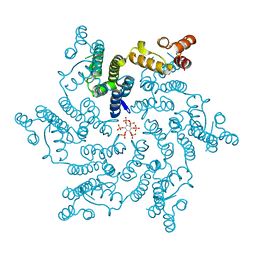 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag protein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2017-10-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
6ERN
 
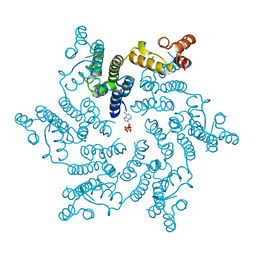 | | HIV Hexamer with ligand | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Gag polyprotein | | Authors: | James, L.C. | | Deposit date: | 2017-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | HIV hexamer
To Be Published
|
|
6ERM
 
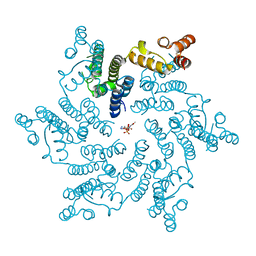 | | HIV Hexamer with ligand | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, Gag polyprotein | | Authors: | James, L.C. | | Deposit date: | 2017-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV hexamer
To Be Published
|
|
8G1N
 
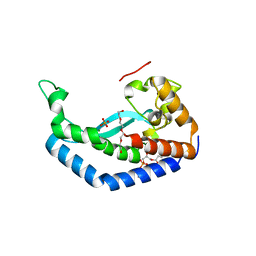 | | Structure of Campylobacter concisus PglC I57M/Q175M Variant with modeled C-terminus | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MAGNESIUM ION, N,N'-diacetylbacilliosaminyl-1-phosphate transferase, ... | | Authors: | Dodge, G.J, Ray, L.C, Das, D, Imperiali, B, Allen, K.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Co-conserved sequence motifs are predictive of substrate specificity in a family of monotopic phosphoglycosyl transferases.
Protein Sci., 32, 2023
|
|
2PFP
 
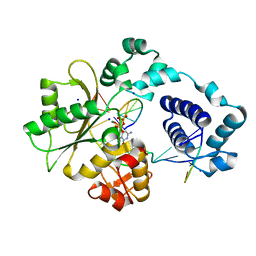 | | DNA Polymerase lambda in complex with DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, Downstream Primer, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
2DSM
 
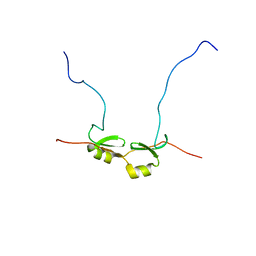 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
2E4G
 
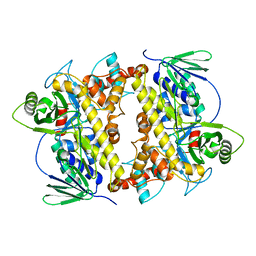 | | RebH with bound L-Trp | | Descriptor: | TRYPTOPHAN, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
6ME2
 
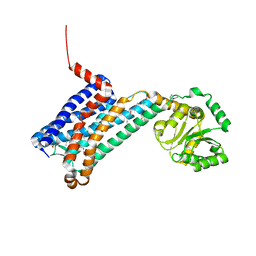 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6ME4
 
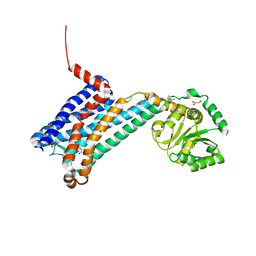 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6M4G
 
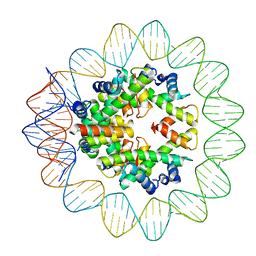 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (93-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
7Q89
 
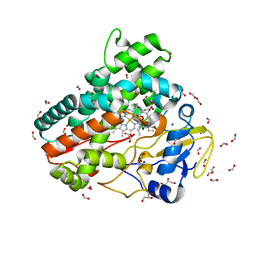 | | OleP mutant G92W in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-10 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7Q6X
 
 | | OleP mutant S240Y in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
