5HOT
 
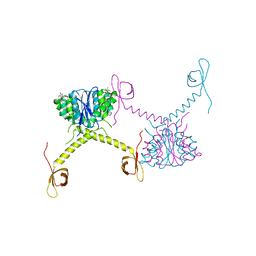 | | Structural Basis for Inhibitor-Induced Aggregation of HIV-1 Integrase | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, Integrase | | Authors: | Gupta, K, Turkki, V, Sherrill-Mix, S, Hwang, Y, Eilers, G, Taylor, L, McDanal, C, Wang, P, Temelkoff, D, Nolte, R, Velthuisen, E, Jeffrey, J, Van Duyne, G.D, Bushman, F.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5HXW
 
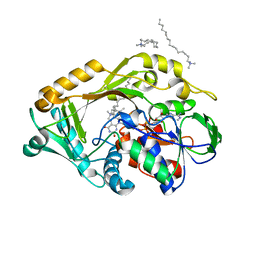 | | L-amino acid deaminase from Proteus vulgaris | | Descriptor: | CETYL-TRIMETHYL-AMMONIUM, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-01-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
5HZY
 
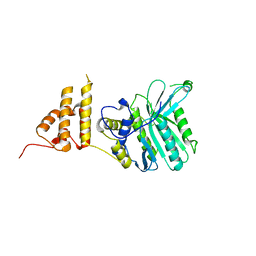 | | Crystal structure of the legionella pneumophila effector protein RavZ - P6322 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-02-03 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5HVN
 
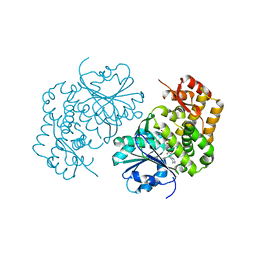 | | 3.0 Angstrom Crystal Structure of 3-dehydroquinate Synthase (AroB) from Francisella tularensis in Complex with NAD. | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Zhou, M, Grimshaw, S, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3.0 Angstrom Crystal Structure of 3-dehydroquinate Synthase (AroB) from Francisella tularensis in Complex with NAD.
To Be Published
|
|
5HSF
 
 | | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1. | | Descriptor: | Ig gamma-1 chain C region, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Flores, K, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1.
To Be Published
|
|
5HPE
 
 | | Phosphatase domain of PP5 bound to a phosphomimetic Cdc37 substrate peptide | | Descriptor: | COBALT HEXAMMINE(III), MANGANESE (II) ION, Serine/threonine-protein phosphatase 5,Hsp90 co-chaperone Cdc37 | | Authors: | Oberoi, J, Mariotti, L, Vaughan, C. | | Deposit date: | 2016-01-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and functional basis of protein phosphatase 5 substrate specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HRJ
 
 | |
190D
 
 | |
5HVW
 
 | | Monomeric IgG4 Fc | | Descriptor: | GLYCEROL, Ig gamma-4 chain C region, ZINC ION, ... | | Authors: | Oganesyan, V.Y, Shan, L, Dall'Acqua, W.F. | | Deposit date: | 2016-01-28 | | Release date: | 2016-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Generation and Characterization of an IgG4 Monomeric Fc Platform.
Plos One, 11, 2016
|
|
5I2H
 
 | | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, FORMIC ACID, ... | | Authors: | Chang, C, Duke, N, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-08 | | Release date: | 2016-03-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin.
To Be Published
|
|
5I39
 
 | | High resolution structure of L-amino acid deaminase from Proteus vulgaris with the deletion of the specific insertion sequence | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
5I2U
 
 | | Crystal structure of a novel Halo-Tolerant Cellulase from Soil Metagenome | | Descriptor: | Cellulase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garg, R, Brahma, V, Srivastava, R, Verma, L, Karthikeyan, S, Sahni, G. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome
Sci Rep, 6, 2016
|
|
5I45
 
 | | 1.35 Angstrom Crystal Structure of C-terminal Domain of Glycosyl Transferase Group 1 Family Protein (LpcC) from Francisella tularensis. | | Descriptor: | Glycosyl transferases group 1 family protein | | Authors: | Minasov, G, Filippova, E, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-11 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 1.35 Angstrom Crystal Structure of C-terminal Domain of Glycosyl Transferase Group 1 Family Protein (LpcC) from Francisella tularensis.
To Be Published
|
|
5I23
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF022 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-{[(1S,2R,3S,4S,5R,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)cyclohexyl]amino}octyl)triaza-1,2-dien-2-ium, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
192D
 
 | | RECOMBINATION-LIKE STRUCTURE OF D(CCGCGG) | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*GP*G)-3'), SODIUM ION | | Authors: | Malinina, L, Urpi, L, Salas, X, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1994-09-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Recombination-like structure of d(CCGCGG).
J.Mol.Biol., 243, 1994
|
|
1A3I
 
 | | X-RAY CRYSTALLOGRAPHIC DETERMINATION OF A COLLAGEN-LIKE PEPTIDE WITH THE REPEATING SEQUENCE (PRO-PRO-GLY) | | Descriptor: | ACETIC ACID, COLLAGEN-LIKE PEPTIDE | | Authors: | Kramer, R.Z, Vitagliano, L, Bella, J, Berisio, R, Mazzarella, L, Brodsky, B, Zagari, A, Berman, H.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic determination of a collagen-like peptide with the repeating sequence (Pro-Pro-Gly).
J.Mol.Biol., 280, 1998
|
|
1A3J
 
 | | X-RAY CRYSTALLOGRAPHIC DETERMINATION OF A COLLAGEN-LIKE PEPTIDE WITH THE REPEATING SEQUENCE (PRO-PRO-GLY) | | Descriptor: | COLLAGEN-LIKE PEPTIDE | | Authors: | Kramer, R.Z, Vitagliano, L, Bella, J, Berisio, R, Mazzarella, L, Brodsky, B, Zagari, A, Berman, H.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-05-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic determination of a collagen-like peptide with the repeating sequence (Pro-Pro-Gly).
J.Mol.Biol., 280, 1998
|
|
6TAV
 
 | | Crystal structure of endopeptidase-induced alpha2-macroglobulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Gomis-Ruth, F.X, Duquerroy, S, Trapani, S, Marrero, A, Goulas, T, Guevara, T, Andersen, G.A, Sottrup-Jensen, L, Navaza, J. | | Deposit date: | 2019-10-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Cryo-EM structures shows the mechanistic basis of pan-peptidase inhibition by human alpha2-macroglobulin
Proc.Natl.Acad.Sci.USA, 2022
|
|
7LVY
 
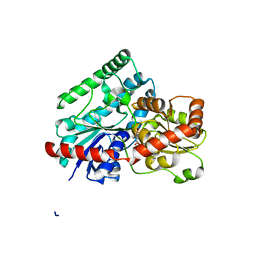 | | Crystal Structure of Tetur04g02350 | | Descriptor: | UDP-glycosyltransferase 203A2, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose | | Authors: | Danehsian, L, Kluza, A, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2021-02-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Tetur04g02350
To Be Published
|
|
7LVM
 
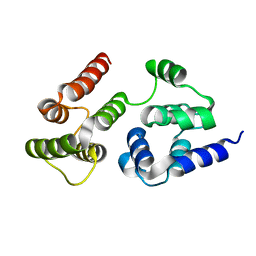 | |
7LVJ
 
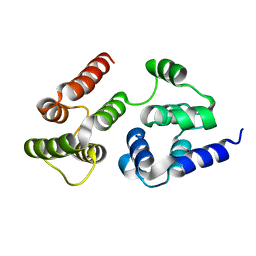 | | CASP8 isoform G DED domain | | Descriptor: | Isoform 9 of Caspase-8 | | Authors: | Weichert, K, Lu, F, Kodandapani, L, Sauder, J.M. | | Deposit date: | 2021-02-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caspase-8 Variant G Regulates Rheumatoid Arthritis Fibroblast-Like Synoviocyte Aggressive Behavior.
ACR Open Rheumatol, 4, 2022
|
|
8Y8K
 
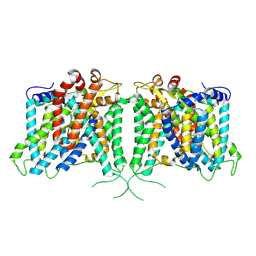 | | The structure of hAE3 | | Descriptor: | Anion exchange protein 3 | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3
Nat Commun, 15, 2024
|
|
8ZLE
 
 | | hAE3NTD2TMD with PT5,CLR, and Y01 | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate, ... | | Authors: | Jian, L, Zhang, Q, Yao, D, Wang, Q, Xia, Y, Qin, A, Cao, Y. | | Deposit date: | 2024-05-19 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3
Nat Commun, 15, 2024
|
|
7LCW
 
 | | Aspartimidylated Lasso Peptide Lihuanodin | | Descriptor: | Lasso Peptide Lihuanodin | | Authors: | Link, A.J, Cao, L. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cellulonodin-2 and Lihuanodin: Lasso Peptides with an Aspartimide Post-Translational Modification.
J.Am.Chem.Soc., 143, 2021
|
|
7LP4
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
