2PLW
 
 | | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052). | | Descriptor: | Ribosomal RNA methyltransferase, putative, S-ADENOSYLMETHIONINE, ... | | Authors: | Wernimont, A.K, Hassanali, A, Lin, L, Lew, J, Zhao, Y, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052).
To be Published
|
|
6LM2
 
 | |
2PNC
 
 | | Crystal Structure of Bovine Plasma Copper-Containing Amine Oxidase in Complex with Clonidine | | Descriptor: | 2,6-DICHLORO-N-IMIDAZOLIDIN-2-YLIDENEANILINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Cendron, L, Holt, A, Smith, D.J, Zanotti, G, Rigo, A, Di Paolo, M.L. | | Deposit date: | 2007-04-24 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple binding sites for substrates and modulators of semicarbazide-sensitive amine oxidases: kinetic consequences
Mol.Pharmacol., 73, 2008
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
2PVQ
 
 | | Crystal structure of Ochrobactrum anthropi glutathione transferase Cys10Ala mutant with glutathione bound at the H-site | | Descriptor: | GLUTATHIONE, SULFATE ION, glutathione S-transferase | | Authors: | Allocati, N, Federici, L, Masulli, M, Favaloro, B, Di Ilio, C. | | Deposit date: | 2007-05-10 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cysteine 10 is critical for the activity of Ochrobactrum anthropi glutathione transferase and its mutation to alanine causes the preferential binding of glutathione to the H-site.
Proteins, 71, 2008
|
|
6LPC
 
 | | Crystal Structure of rat Munc18-1 with K332E/K333E mutation | | Descriptor: | Syntaxin-binding protein 1 | | Authors: | Wang, X.P, Gong, J.H, Wang, S, Zhu, L, Yang, X.Y, Xu, Y.Y, Yang, X.F, Ma, C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly.
Embo J., 39, 2020
|
|
6LPS
 
 | |
2Q19
 
 | | 2-keto-3-deoxy-D-arabinonate dehydratase apo form | | Descriptor: | 2-keto-3-deoxy-D-arabinonate dehydratase | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
4NPQ
 
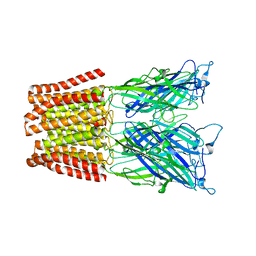 | | The resting-state conformation of the GLIC ligand-gated ion channel | | Descriptor: | Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2Q1C
 
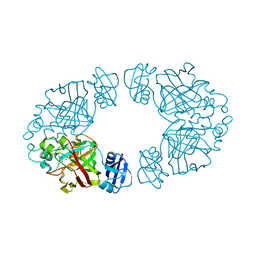 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with calcium and 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, CALCIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
2Q2G
 
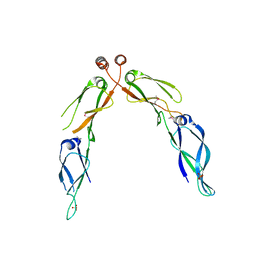 | | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800 | | Descriptor: | Heat shock 40 kDa protein, putative (fragment), SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Lin, L, Hassanali, A, Kozieradzki, I, Wasney, G, Vedadi, M, Walker, J.R, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800.
To be Published
|
|
4NBY
 
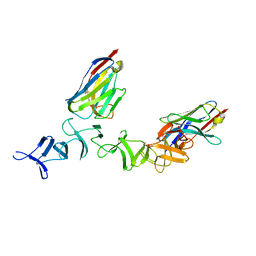 | | Crystal Structure of TcdA-A2 Bound to Two Molecules of A20.1 VHH | | Descriptor: | A20.1 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
6LT9
 
 | |
2PH1
 
 | | Crystal structure of nucleotide-binding protein AF2382 from Archaeoglobus fulgidus, Northeast Structural Genomics Target GR165 | | Descriptor: | Nucleotide-binding protein, ZINC ION | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-04-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of nucleotide-binding protein AF2382 from Archaeoglobus fulgidus.
To be Published
|
|
4NC1
 
 | | Crystal Structure of TcdA-A2 Bound to A20.1 VHH and A26.8 VHH | | Descriptor: | A20.1 VHH, A26.8 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
2PSB
 
 | | Crystal structure of YerB protein from Bacillus subtilis. NorthEast Structural Genomics target SR586 | | Descriptor: | YerB protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-04 | | Release date: | 2007-05-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YerB protein from Bacillus subtilis.
To be Published
|
|
4NED
 
 | | Crystal STRUCTURE OF C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH FENOPROFEN AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Gautam, L, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-29 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal STRUCTURE OF C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH FENOPROFEN AT 2.1 ANGSTROM RESOLUTION
To be Published
|
|
2PT7
 
 | | Crystal structure of Cag VirB11 (HP0525) and an inhibitory protein (HP1451) | | Descriptor: | Cag-alfa, Hypothetical protein | | Authors: | Hare, S, Fischer, W, Williams, R, Terradot, L, Bayliss, R, Haas, R, Waksman, G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase.
Embo J., 26, 2007
|
|
6M17
 
 | | The 2019-nCoV RBD/ACE2-B0AT1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2020-02-24 | | Release date: | 2020-03-11 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
2PNU
 
 | | Crystal structure of human androgen receptor ligand-binding domain in complex with EM-5744 | | Descriptor: | (5S,8R,9S,10S,13R,14S,17S)-13-{2-[(3,5-DIFLUOROBENZYL)OXY]ETHYL}-17-HYDROXY-10-METHYLHEXADECAHYDRO-3H-CYCLOPENTA[A]PHENANTHREN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Cantin, L, Faucher, F, Couture, J.F, Pereira de Jesus-Tran, K, Legrand, P, Ciobanu, C.L, Singh, S.M, Labrie, F, Breton, R. | | Deposit date: | 2007-04-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Androgen Receptor Ligand-binding Domain Complexed with EM5744, a Rationally Designed Steroidal Ligand Bearing a Bulky Chain Directed toward Helix 12.
J.Biol.Chem., 282, 2007
|
|
2Q2M
 
 | |
4NM6
 
 | | Crystal structure of TET2-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*T)-3', FE (II) ION, Methylcytosine dioxygenase TET2, ... | | Authors: | Hu, L, Li, Z, Cheng, J, Rao, Q, Gong, W, Liu, M, Wang, P, Xu, Y. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation.
Cell(Cambridge,Mass.), 155, 2013
|
|
6LQI
 
 | | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Geng, J, Liu, W, Zhou, H, Zhang, T, Wang, L, Zhang, M, Shen, B, Li, X, Xiao, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel.
Neuron, 106, 2020
|
|
4NNP
 
 | |
2PG0
 
 | | Crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus | | Descriptor: | Acyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Li, Y, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acyl-CoA dehydrogenase from G. kaustophilus
To be Published
|
|
