1GSD
 
 | |
1GSF
 
 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
3ZPP
 
 | |
4V1N
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4UUL
 
 | |
4UWI
 
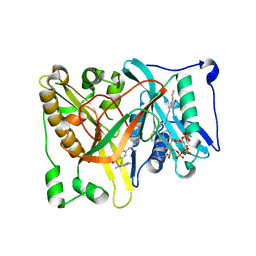 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-4-[3-(4-methylpiperazin-1-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
4W93
 
 | | Human pancreatic alpha-amylase in complex with montbretin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Montbretin A, ... | | Authors: | Williams, L.K, Caner, S, Brayer, G.D. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif.
Nat.Chem.Biol., 11, 2015
|
|
4UWQ
 
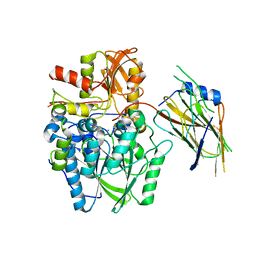 | | Crystal structure of the disulfide-linked complex of the thiosulfodyrolase SoxB with the carrier-protein SoxYZ from Thermus thermophilus | | Descriptor: | MANGANESE (II) ION, SOXY PROTEIN, SOXZ, ... | | Authors: | Grabarczyk, D.B, Chappell, P.E, Johnson, S, Stelzl, L.S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-08-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Basis for Specificity and Promiscuity in a Carrier Protein/Enzyme System from the Sulfur Cycle
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V99
 
 | |
4V4Z
 
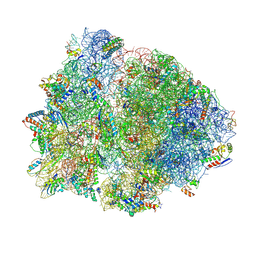 | | 70S Thermus thermophilous ribosome functional complex with mRNA and E- and P-site tRNAs at 4.5A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4V9J
 
 | | 70S ribosome translocation intermediate GDPNP-II containing elongation factor EFG/GDPNP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.86 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
4V25
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-chloro-5-methylpyrimidin-4-yl)phenyl]-N-(4-{[(difluoroacetyl)amino]methyl}benzyl)-2,4-dihydroxybenzamide, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4V72
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4UVW
 
 | | Crystal structure of human tankyrase 2 in complex with 4,5-dimethyl-3- phenyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 4,5-dimethyl-3-phenylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4V26
 
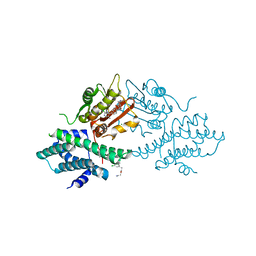 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-CHLORO-5-METHYLPYRIMIDIN-4-YL)PHENYL]-2,4-DIHYDROXY-N-(4-{[(TRIFLUOROACETYL)AMINO]METHYL}BENZYL)BENZAMIDE, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4V88
 
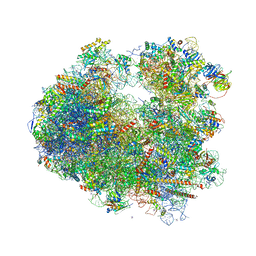 | | The structure of the eukaryotic ribosome at 3.0 A resolution. | | Descriptor: | 18S RIBOSOMAL RNA, 18S rRNA, 25S rRNA, ... | | Authors: | Ben-Shem, A, Garreau de Loubresse, N, Melnikov, S, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2011-10-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the eukaryotic ribosome at 3.0 angstrom resolution.
Science, 334, 2011
|
|
4UX4
 
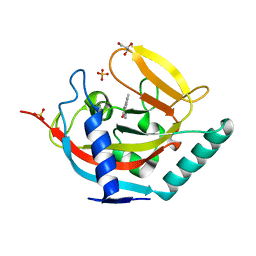 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-(4- methylphenyl)-5-oxo-5,6-dihydro-1,6-naphthyridin-1-ium | | Descriptor: | (1S)-1-methyl-7-(4-methylphenyl)-5-oxo-1,5-dihydro-1,6-naphthyridin-1-ium, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-19 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis and Evaluation in Vitro of Arylnaphthyridinones, Arylpyridopyrimidinones and Their Tetrahydro Derivatives as Inhibitors of the Tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
4WA2
 
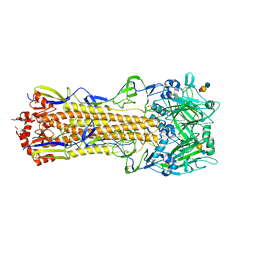 | | The crystal structure of hemagglutinin from a H3N8 influenza virus isolated from New England harbor seals in complex with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
6ZT9
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT7
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
4W8A
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4UQM
 
 | | Crystal structure determination of uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans in complex with DNA - new insights into the role of the Leucine-loop for damage recognition and repair | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*CP*CP*AP*AAB*GP*TP*CP*TP*CP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Pedersen, H.L, Johnson, K.A, McVey, C.E, Leiros, I, Moe, E. | | Deposit date: | 2014-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure determination of uracil-DNA N-glycosylase from Deinococcus radiodurans in complex with DNA.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4UQJ
 
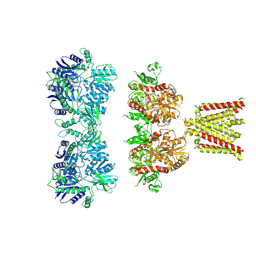 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UUP
 
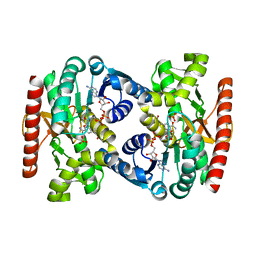 | | Reconstructed ancestral trichomonad malate dehydrogenase in complex with NADH, SO4, and PO4 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Steindel, P.A, Chen, E.H, Theobald, D.L. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Gradual Neofunctionalization in the Convergent Evolution of Trichomonad Lactate and Malate Dehydrogenases.
Protein Sci., 25, 2016
|
|
4UYJ
 
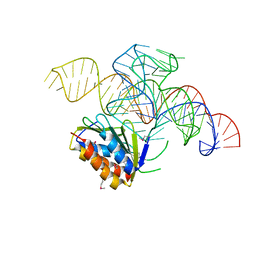 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
