4DE5
 
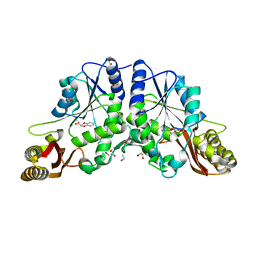 | | Pantothenate synthetase in complex with fragment 6 | | Descriptor: | (2S)-2,3-dihydro-1,4-benzodioxine-2-carboxylic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L. | | Deposit date: | 2012-01-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5XSA
 
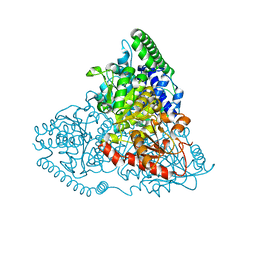 | | Crystal Structure of Transketolase in complex with TPP intermediate III from Pichia Stipitis | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, Transketolase | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-13 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.975 Å) | | Cite: | The Mesomeric Effect of Thiazolium on non-Kekule Diradicals in Pichia stipitis Transketolase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XTX
 
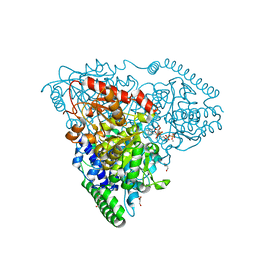 | | Crystal Structure of Transketolase in complex with TPP intermediate VII from Pichia Stipitis | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methylidene-2-[(2S,3S,4R,5R)-1,2,3,4,5-pentakis(oxidanyl)-6-phosphonooxy-hexan-2-yl]-1,3-thiazolidin-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.049 Å) | | Cite: | Evidence of Diradicals Involved in the Yeast Transketolase Catalyzed Keto-Transferring Reactions.
Chembiochem, 19, 2018
|
|
4DDM
 
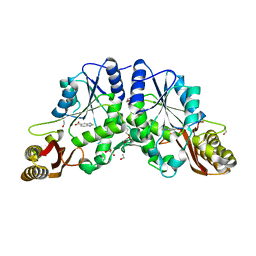 | | Pantothenate synthetase in complex with 2,1,3-benzothiadiazole-5-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2,1,3-benzothiadiazole-5-carboxylic acid, ETHANOL, ... | | Authors: | Silvestre, H.L. | | Deposit date: | 2012-01-18 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2P54
 
 | | a crystal structure of PPAR alpha bound with SRC1 peptide and GW735 | | Descriptor: | 2-METHYL-2-(4-{[({4-METHYL-2-[4-(TRIFLUOROMETHYL)PHENYL]-1,3-THIAZOL-5-YL}CARBONYL)AMINO]METHYL}PHENOXY)PROPANOIC ACID, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Xu, R.X, Xu, H.E, Sierra, M.L, Montana, V.G, Lambert, M.H, Pianetti, P.M. | | Deposit date: | 2007-03-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substituted 2-[(4-Aminomethyl)phenoxy]-2-methylpropionic Acid PPAR Agonists. 1.Discovery of a Novel Series of Potent HDLc Raising Agents.
J.Med.Chem., 50, 2007
|
|
5XVL
 
 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
3AQ0
 
 | | Ligand-bound form of Arabidopsis medium/long-chain length prenyl pyrophosphate synthase (surface polar residue mutant) | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, DI(HYDROXYETHYL)ETHER, FARNESYL DIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase
Plant Physiol., 155, 2011
|
|
1KW5
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-01-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
6ZE5
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE6
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 4-nitrocatechol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITROCATECHOL, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1U7D
 
 | |
6ZE7
 
 | | Chaetomium thermophilum FAD-dependent oxidoreductase in complex with 4-nitrophenol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3AU0
 
 | | Structural and biochemical characterization of ClfB:ligand interactions | | Descriptor: | Clumping factor B, MAGNESIUM ION | | Authors: | Ganesh, V.K, Barbu, E.M, Deivanayagam, C.C.S, Le, B, Anderson, A.S, Matsuka, Y, Lin, S.L, Foster, T.F, Narayana, S.V.L, Hook, M. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biochemical characterization of ClfB:ligand interactions
To be published
|
|
4DBP
 
 | |
5XPS
 
 | | Crystal Structure of Transketolase in complex with erythrose-4-phosphate from Pichia Stipitis | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, ERYTHOSE-4-PHOSPHATE, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-04 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Evidence of Diradicals Involved in the Yeast Transketolase Catalyzed Keto-Transferring Reactions.
Chembiochem, 19, 2018
|
|
6EYL
 
 | | Crystal structure of OpuBC in complex with carnitine | | Descriptor: | CARNITINE, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Peherstorfer, S, Teichmann, L, Smits, S.H, Sschmitt, L, Bremer, E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reprogramming the substrate specificity of an ABC import system by a single amino acid substitution in its cognate ligand binding protein
To Be Published
|
|
5XSM
 
 | | Crystal Structure of Transketolase in complex with TPP intermediate IV from Pichia Stipitis | | Descriptor: | 2-[(5S)-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2,5-dihydro-1,3-thiazol-3-ium-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Mesomeric Effect of Thiazolium on non-Kekule Diradicals in Pichia stipitis Transketolase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1KGT
 
 | | Crystal Structure of Tetrahydrodipicolinate N-Succinyltransferase in Complex with Pimelate and Succinyl-CoA | | Descriptor: | 2,3,4,5-TETRAHYDROPYRIDINE-2-CARBOXYLATE N-SUCCINYLTRANSFERASE, PIMELIC ACID, SUCCINYL-COENZYME A | | Authors: | Beaman, T.W, Vogel, K.W, Drueckhammer, D.G, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2001-11-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acyl group specificity at the active site of tetrahydridipicolinate N-succinyltransferase.
Protein Sci., 11, 2002
|
|
2P2V
 
 | | Crystal structure analysis of monofunctional alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase, CHLORIDE ION, ... | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
2P4E
 
 | | Crystal Structure of PCSK9 | | Descriptor: | MERCURY (II) ION, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Cunningham, D, Danley, D.E, Geoghegan, F.K, Griffor, M.C, Hawkins, J.L, Qiu, X. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8QNO
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase treated at 368 K from Pyrococcus furiosus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Koeppl, L.H, Popadic, D, Andexer, J.N. | | Deposit date: | 2023-09-27 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
7T4S
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4Q
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
1TN8
 
 | | Protein Farnesyltransferase Complexed with a H-Ras Peptide Substrate and a FPP Analog at 2.25A Resolution | | Descriptor: | ACETIC ACID, Protein farnesyltransferase beta subunit, Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
