3R0M
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 | | Descriptor: | Llama VHH A12, SULFATE ION | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-03-08 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Single-Headed Immunoglobulins Efficiently Penetrate CD4-Binding Site and Effectively Neutralize HIV-1
To be Published
|
|
3PIQ
 
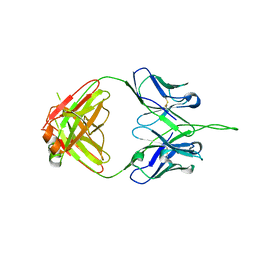 | | Crystal structure of human 2909 Fab, a quaternary structure-specific antibody against HIV-1 | | Descriptor: | Human monoclonal antibody 2909 Fab heavy chain, Human monoclonal antibody 2909 Fab light chain | | Authors: | Changela, A, Gorny, M.K, Zolla-Pazner, S, Kwong, P.D. | | Deposit date: | 2010-11-07 | | Release date: | 2011-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.325 Å) | | Cite: | Crystal Structure of Human Antibody 2909 Reveals Conserved Features of Quaternary Structure-Specific Antibodies That Potently Neutralize HIV-1.
J.Virol., 85, 2011
|
|
3Q39
 
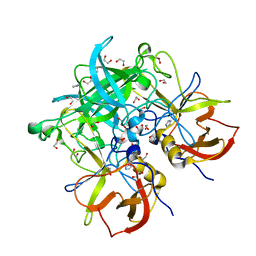 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with HBGA type H2 (diglycan) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE, ... | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
3RPT
 
 | |
3RJQ
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 in complex with C186 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C186 gp120, Llama VHH A12 | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of anti-HIV A12 VHH of llama antibody in complex with C1086 gp120
To be Published
|
|
3R6K
 
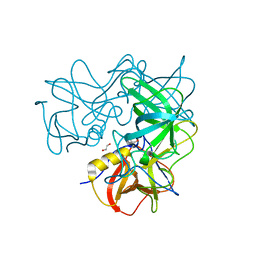 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 in complex with HBGA type B (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
3RRT
 
 | |
3RRR
 
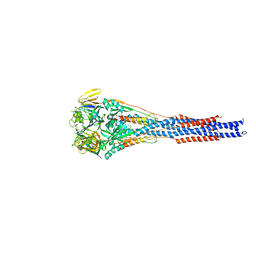 | | Structure of the RSV F protein in the post-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | McLellan, J.S, Yongping, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2011-04-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes.
J.Virol., 85, 2011
|
|
3RY8
 
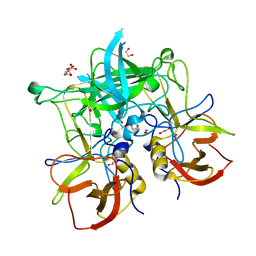 | | Structural basis for norovirus inhibition and fucose mimicry by citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Capsid protein | | Authors: | Hansman, G.S, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-05-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for norovirus inhibition and fucose mimicry by citrate.
J.Virol., 86, 2012
|
|
3PA1
 
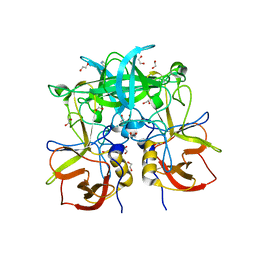 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with HBGA type A | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE, ... | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-10-18 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structures of GII.10 and GII.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability.
J.Virol., 85, 2011
|
|
7KHF
 
 | |
7KFK
 
 | |
7KJP
 
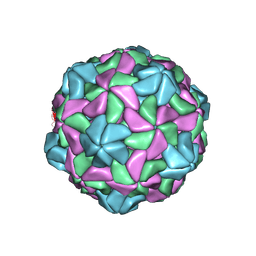 | | Disulfide Stabilized Norovirus GI.1 VLP Shell Region | | Descriptor: | Capsid protein VP1 | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Disulfide stabilization of human norovirus GI.1 virus-like particles focuses immune response toward blockade epitopes.
NPJ Vaccines, 5, 2020
|
|
4ZPT
 
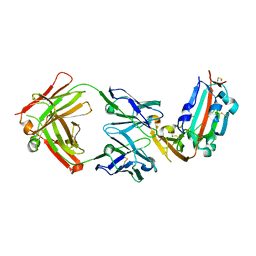 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV
Nat Commun, 6, 2015
|
|
7KC1
 
 | |
7JZ4
 
 | |
7JZK
 
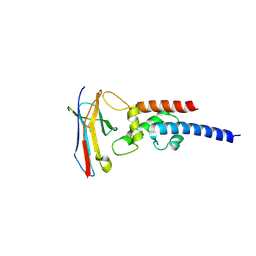 | |
7JZ1
 
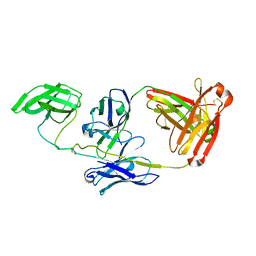 | |
7JZI
 
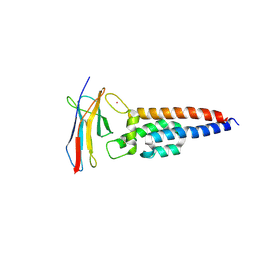 | |
7JKS
 
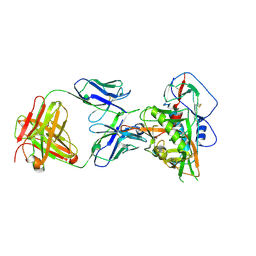 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7KNI
 
 | | Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNE
 
 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KMB
 
 | | ACE2-RBD Focused Refinement Using Symmetry Expansion of Applied C3 for Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNH
 
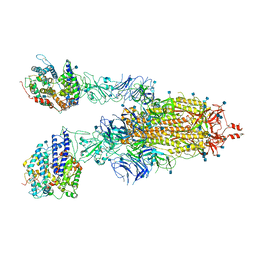 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
