4XIG
 
 | | Crystal structure of bacterial alginate ABC transporter determined through humid air and glue-coating method | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
3CE1
 
 | | Crystal Structure of the Cu/Zn Superoxide Dismutase from Cryptococcus liquefaciens Strain N6 | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Teh, A.H, Kanamasa, S, Kajiwara, S, Kumasaka, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Cu/Zn superoxide dismutase from the heavy-metal-tolerant yeast Cryptococcus liquefaciens strain N6.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
4XTC
 
 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | Descriptor: | AlgM1, AlgM2, AlgQ2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
7QY4
 
 | | As isolated MSOX movie series dataset 5 (2 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QYC
 
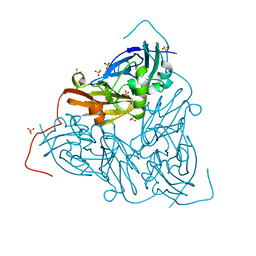 | | As isolated MSOX movie series dataset 20 (8 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8JOG
 
 | | solution structure of Ras Binding Domein (RBD) in C-RAF with negative allosteric modulator. | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
8JOF
 
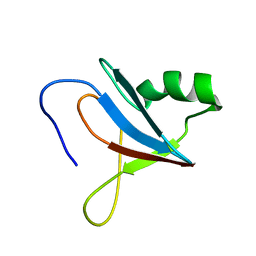 | | solution-structure of Ras Binding Domain (RBD) in C-RAF | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
7WB8
 
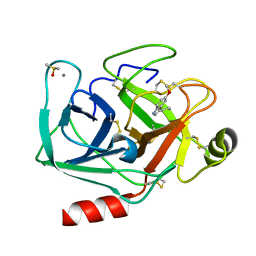 | | Crystal structure of Bovine Pancreatic Trypsin in complex with 5-Methoxytryptamine at Room Temperature | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Sakai, N, Okumura, H, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | In situ crystal data-collection and ligand-screening system at SPring-8.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7WB6
 
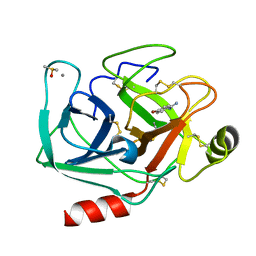 | | Crystal structure of Bovine Pancreatic Trypsin in complex with 4-Bromobenzamidine at Room Temperature | | Descriptor: | 4-bromanylbenzenecarboximidamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Sakai, N, Okumura, H, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | In situ crystal data-collection and ligand-screening system at SPring-8.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7WB9
 
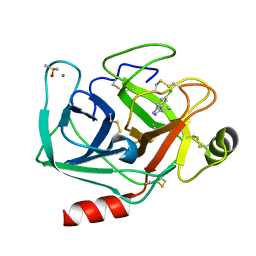 | | Crystal structure of Bovine Pancreatic Trypsin in complex with 5-Chlorotryptamine at Room Temperature | | Descriptor: | 2-(5-chloranyl-1~{H}-indol-3-yl)ethanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Sakai, N, Okumura, H, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | In situ crystal data-collection and ligand-screening system at SPring-8.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7WA0
 
 | | Crystal structure of Bovine Pancreatic Trypsin in complex with Benzamidine at Room Temperature | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Sakai, N, Okumura, H, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2021-12-11 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | In situ crystal data-collection and ligand-screening system at SPring-8.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7WBA
 
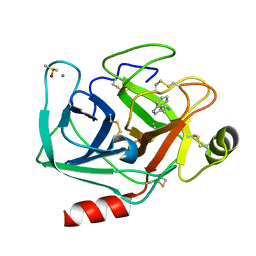 | | Crystal structure of Bovine Pancreatic Trypsin in complex with Tryptamine at Room Temperature | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Sakai, N, Okumura, H, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | In situ crystal data-collection and ligand-screening system at SPring-8.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7WA2
 
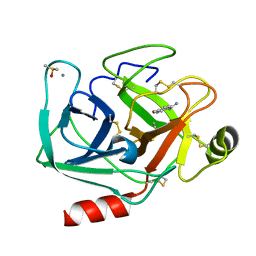 | | Crystal structure of Bovine Pancreatic Trypsin in complex with 4-Methoxybenzamidine at Room Temperature | | Descriptor: | 4-methoxybenzenecarboximidamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Sakai, N, Okumura, H, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2021-12-11 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | In situ crystal data-collection and ligand-screening system at SPring-8.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7WB7
 
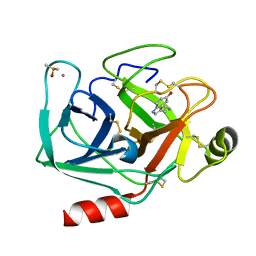 | | Crystal structure of Bovine Pancreatic Trypsin in complex with Serotonin at Room Temperature | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Sakai, N, Okumura, H, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | In situ crystal data-collection and ligand-screening system at SPring-8.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | Descriptor: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | Authors: | Fibriansah, G, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
4EFL
 
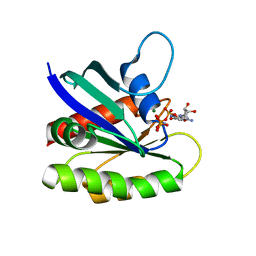 | | Crystal structure of H-Ras WT in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
4EFN
 
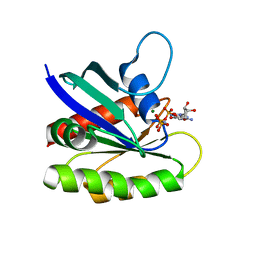 | | Crystal structure of H-Ras Q61L in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
4EFM
 
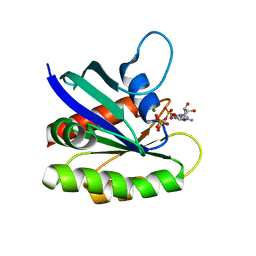 | | Crystal structure of H-Ras G12V in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
5B42
 
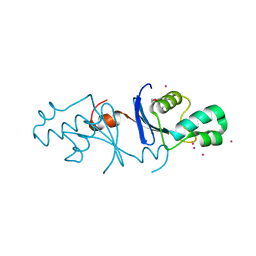 | | Crystal structure of the C-terminal endonuclease domain of Aquifex aeolicus MutL. | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL | | Authors: | Fukui, K, Baba, S, Kumasaka, T, Yano, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Features and Functional Dependency on beta-Clamp Define Distinct Subfamilies of Bacterial Mismatch Repair Endonuclease MutL
J.Biol.Chem., 291, 2016
|
|
2GRU
 
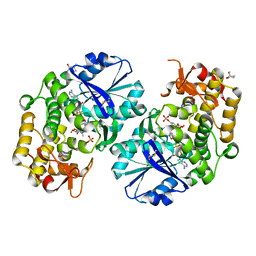 | | Crystal structure of 2-deoxy-scyllo-inosose synthase complexed with carbaglucose-6-phosphate, NAD+ and Co2+ | | Descriptor: | (1R,2S,3S,4R)-5-METHYLENECYCLOHEXANE-1,2,3,4-TETRAOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-deoxy-scyllo-inosose synthase, ... | | Authors: | Nango, E, Kumasaka, T. | | Deposit date: | 2006-04-25 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+
Proteins, 70, 2008
|
|
7YVZ
 
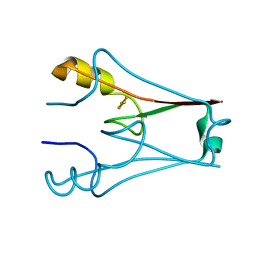 | | Structure of Caenorhabditis elegans CISD-1/mitoNEET | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1 (CISD-1/mitoNEET), FE2/S2 (INORGANIC) CLUSTER | | Authors: | Hasegawa, K, Hagiuda, E, Taguchi, A.T, Geldenhuys, W, Iwasaki, T, Kumasaka, T. | | Deposit date: | 2022-08-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and biological evaluation of Caenorhabditis elegans CISD-1/mitoNEET, a KLP-17 tail domain homologue, supports attenuation of paraquat-induced oxidative stress through a p38 MAPK-mediated antioxidant defense response.
Adv Redox Res, 6, 2022
|
|
7ZCS
 
 | | Nitrite-bound MSOX movie series dataset 65 (52 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - water ligand (final) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCP
 
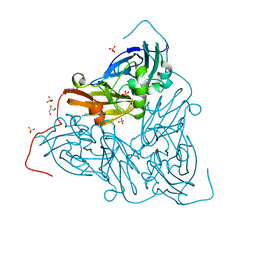 | | Nitrite-bound MSOX movie series dataset 17 (13.6 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitric oxide (NO) intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCO
 
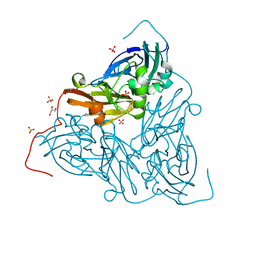 | | Nitrite-bound MSOX movie series dataset 8 (6.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - nitrite/NO intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCQ
 
 | | Nitrite-bound MSOX movie series dataset 25 (20 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) - NO/water intermediate | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-03-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
