4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | Authors: | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.357 Å) | | Cite: | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
2F33
 
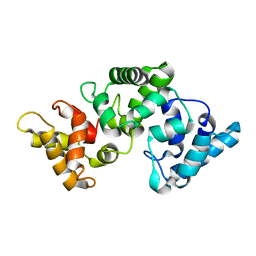 | | NMR solution structure of Ca2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
2G9B
 
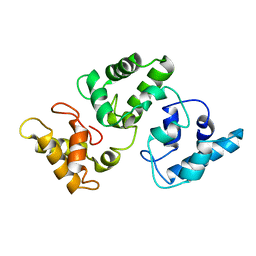 | | NMR solution structure of CA2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2006-03-06 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
6AO9
 
 | |
6AOA
 
 | |
6AOB
 
 | |
2HD4
 
 | | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution | | Descriptor: | 8-mer Peptide from Lactotransferrin, ACETIC ACID, CALCIUM ION, ... | | Authors: | Prem Kumar, R, Singh, A.K, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-20 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution
To be Published
|
|
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
4E6K
 
 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | Descriptor: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
2DVC
 
 | | Structure of the bovine lactoferrin C-lobe complex with sucrose at 3.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem Kumar, R, Bhardwaj, R, Ethayathulla, A.S, Sinha, M, Singh, N, Bhushan, A, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-07-31 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the bovine lactoferrin C-lobe complex with sucrose at 3.0 A resolution
To be Published
|
|
3MWN
 
 | | Structure of the Novel 14 kDa Fragment of alpha-Subunit of Phycoerythrin from the Starving Cyanobacterium Phormidium Tenue | | Descriptor: | PHYCOCYANOBILIN, PHYCOERYTHRIN | | Authors: | Soni, B.R, Hasan, M.I, Parmar, A, Ethayathulla, A.S, Kumar, R.P, Singh, N.K, Sinha, M, Kaur, P, Yadav, S, Sharma, S, Madamwar, D, Singh, T.P. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the novel 14kDa fragment of alpha-subunit of phycoerythrin from the starving cyanobacterium Phormidium tenue.
J.Struct.Biol., 171, 2010
|
|
8B9R
 
 | | Molecular structure of Cu(II)-bound amyloid-beta monomer implicated in inhibition of peptide self-assembly in Alzheimer's disease | | Descriptor: | Amyloid-beta A4 protein, COPPER (II) ION | | Authors: | Abelein, A, Ciofi-Baffoni, S, Morman, C, Kumar, R, Giachetti, A, Piccioli, M, Biverstal, H. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Structure of Cu(II)-Bound Amyloid-beta Monomer Implicated in Inhibition of Peptide Self-Assembly in Alzheimer's Disease.
Jacs Au, 2, 2022
|
|
8B9Q
 
 | | Molecular structure of Cu(II)-bound amyloid-beta monomer implicated in inhibition of peptide self-assembly in Alzheimer's disease | | Descriptor: | Amyloid-beta A4 protein, COPPER (II) ION | | Authors: | Abelein, A, Ciofi-Baffoni, S, Kumar, R, Giachetti, A, Piccioli, M, Biverstal, H. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-08 | | Method: | SOLUTION NMR | | Cite: | Molecular Structure of Cu(II)-Bound Amyloid-beta Monomer Implicated in Inhibition of Peptide Self-Assembly in Alzheimer's Disease.
Jacs Au, 2, 2022
|
|
1P8V
 
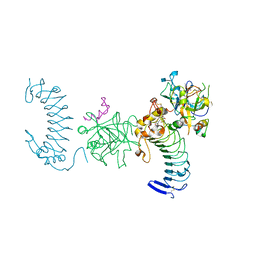 | | CRYSTAL STRUCTURE OF THE COMPLEX OF PLATELET RECEPTOR GPIB-ALPHA AND ALPHA-THROMBIN AT 2.6A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIISOPROPYL PHOSPHONATE, ... | | Authors: | Dumas, J.J, Kumar, R, Seehra, J, Somers, W.S, Mosyak, L. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the GpIbalpha-Thrombin Complex Essential for Platelet Aggregation
Science, 301, 2003
|
|
4E3R
 
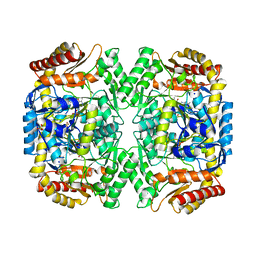 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
3RUV
 
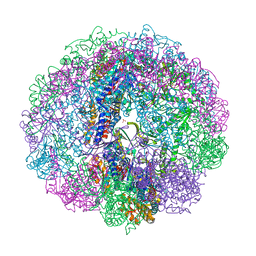 | | Crystal structure of Cpn-rls in complex with ATP analogue from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUS
 
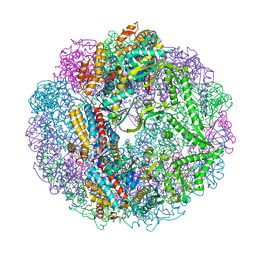 | | Crystal structure of Cpn-rls in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUW
 
 | | Crystal structure of Cpn-rls in complex with ADP-AlFx from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Chaperonin, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
1I5W
 
 | | A-DNA DECAMER GCGTA(TLN)ACGC | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(TLN)P*AP*CP*GP*C)-3' | | Authors: | Egli, M, Minasov, G, Teplova, M, Kumar, R, Wengel, J. | | Deposit date: | 2001-03-01 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of a locked nucleic acid (LNA) duplex composed of a palindromic 10-mer DNA strand containing one LNA thymine monomer
J.Chem.Soc.,Chem.Commun., 2001
|
|
2G58
 
 | | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution | | Descriptor: | (PHQ)IARS, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Prem Kumar, R, Singh, N, Somvanshi, R.K, Ethayathulla, A.S, Dey, S, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution
To be Published
|
|
4M2P
 
 | |
4M2Q
 
 | | Crystal structure of non-myristoylated recoverin with Cysteine-39 oxidized to sulfenic acid | | Descriptor: | CALCIUM ION, Recoverin | | Authors: | Prem Kumar, R, Chakrabarti, K, Kern, D, Oprian, D.D. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Highly Conserved Cysteine of Neuronal Calcium-sensing Proteins Controls Cooperative Binding of Ca2+ to Recoverin.
J.Biol.Chem., 288, 2013
|
|
4M2O
 
 | |
3IZJ
 
 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZL
 
 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
