1G8K
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
1G8J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
1A35
 
 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
2OFZ
 
 | | Ultrahigh Resolution Crystal Structure of RNA Binding Domain of SARS Nucleopcapsid (N Protein) at 1.1 Angstrom Resolution in Monoclinic Form. | | Descriptor: | 1,2-ETHANEDIOL, Nucleocapsid protein | | Authors: | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | Deposit date: | 2007-01-04 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
2OG3
 
 | | structure of the rna binding domain of n protein from SARS coronavirus in cubic crystal form | | Descriptor: | Nucleocapsid protein | | Authors: | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | Deposit date: | 2007-01-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
1A31
 
 | | HUMAN RECONSTITUTED DNA TOPOISOMERASE I IN COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*5IUP*5IU*TP*GP*AP*AP*AP*AP*AP*5IUP*5IUP*5IUP*5IUP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*5IUP*5IUP*5IUP*5IUP*CP*AP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), PROTEIN (TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-27 | | Release date: | 1998-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1C5F
 
 | | CRYSTAL STRUCTURE OF THE CYCLOPHILIN-LIKE DOMAIN FROM BRUGIA MALAYI COMPLEXED WITH CYCLOSPORIN A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 1 | | Authors: | Ellis, P.J, Carlow, C.K.S, Ma, D, Kuhn, P. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Complex of Brugia Malayi Cyclophilin and Cyclosporin A.
Biochemistry, 39, 2000
|
|
2OZK
 
 | | Structure of an N-Terminal Truncated Form of Nendou (NSP15) From SARS-CORONAVIRUS | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | Deposit date: | 2007-02-26 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch.
J.Virol., 81, 2007
|
|
3PYP
 
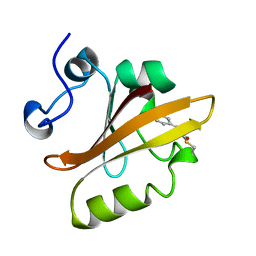 | | PHOTOACTIVE YELLOW PROTEIN, CRYOTRAPPED EARLY LIGHT CYCLE INTERMEDIATE | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Soltis, S.M, Kuhn, P, Canestrelli, I.L, Getzoff, E.D. | | Deposit date: | 1998-07-28 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure at 0.85 A resolution of an early protein photocycle intermediate.
Nature, 392, 1998
|
|
2BBA
 
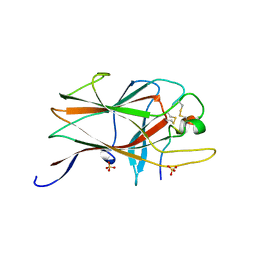 | |
2X60
 
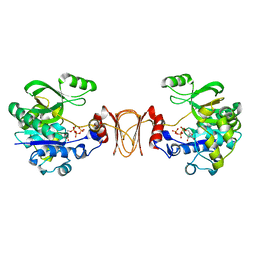 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANNOSE-1-PHOSPHATE GUANYLYLTRANSFERASE | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
2GRI
 
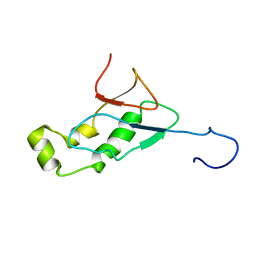 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
4L0P
 
 | | Structure of the human EphA3 receptor ligand binding domain complexed with ephrin-A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Ephrin type-A receptor 3, ... | | Authors: | Forse, G.J, Kolatkar, A.R, Kuhn, P. | | Deposit date: | 2013-05-31 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Distinctive Structure of the EphA3/Ephrin-A5 Complex Reveals a Dual Mode of Eph Receptor Interaction for Ephrin-A5.
Plos One, 10, 2015
|
|
2JZD
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZE
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1O2A
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD at 1.8 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1O27
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and BrdUMP at 2.3 A resolution | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1O29
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and FdUMP at 2.0 A resolution | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
2RNK
 
 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1O26
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and dUMP at 1.6 A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, TRIETHYLENE GLYCOL, ... | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1O2B
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with FAD and PO4 at 2.45 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1O25
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima with dUMP at 2.4 A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
1O24
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima at 2.0 A resolution | | Descriptor: | Thymidylate synthase thyX | | Authors: | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-02-18 | | Release date: | 2003-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
