5XFW
 
 | | Crystal structures of FMN-free form of dihydroorotate dehydrogenase from Trypanosoma brucei | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), MALONATE ION | | Authors: | Kubota, T, Tani, O, Yamaguchi, T, Namatame, I, Sakashita, H, Furukawa, K, Yamasaki, K. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of FMN-bound and FMN-free forms of dihydroorotate dehydrogenase fromTrypanosoma brucei.
FEBS Open Bio, 8, 2018
|
|
5XFV
 
 | | Crystal structures of FMN-bound form of dihydroorotate dehydrogenase from Trypanosoma brucei | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, MALONATE ION | | Authors: | Kubota, T, Tani, O, Yamaguchi, T, Namatame, I, Sakashita, H, Furukawa, K, Yamasaki, K. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of FMN-bound and FMN-free forms of dihydroorotate dehydrogenase fromTrypanosoma brucei.
FEBS Open Bio, 8, 2018
|
|
2D7R
 
 | | Crystal structure of pp-GalNAc-T10 complexed with GalNAc-Ser on lectin domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
2D7I
 
 | | Crystal structure of pp-GalNAc-T10 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
3ASP
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with A-antigen | | Descriptor: | Capsid protein, SODIUM ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3AST
 
 | | Crystal structure of P domain Q389N mutant from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASQ
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with H-antigen | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASR
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-a | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASS
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
8XY5
 
 | |
5CY0
 
 | | Total Chemical Synthesis, Covalent Structure Verification, and X-ray Structure of Bioactive Ts3 Toxin by Racemic Protein Crystallography | | Descriptor: | GLYCEROL, Ts3 Toxin | | Authors: | Dang, B, Kubota, T, Mandal, K, Correa, A.M, Bezanilla, F, Kent, S.B. | | Deposit date: | 2015-07-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Elucidation of the Covalent and Tertiary Structures of Biologically Active Ts3 Toxin.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4LFS
 
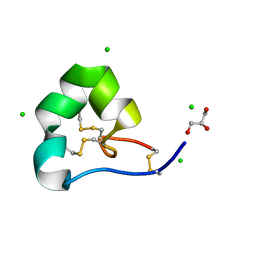 | | High resolution x-ray structure of racemic ShK toxin | | Descriptor: | CHLORIDE ION, GLYCEROL, Potassium channel toxin ShK | | Authors: | Dang, B, Kubota, T, Mandal, K, Bezanilla, F, Kent, S.B.H. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Native Chemical Ligation at Asx-Cys, Glx-Cys: Chemical Synthesis and High-Resolution X-ray Structure of ShK Toxin by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
4LFQ
 
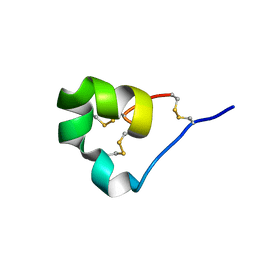 | | High resolution x-ray crystal structure of L-ShK toxin | | Descriptor: | Potassium channel toxin ShK | | Authors: | Dang, B, Kubota, T, Mandal, K, Bezanilla, F, Kent, S.B.H. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.059 Å) | | Cite: | Native Chemical Ligation at Asx-Cys, Glx-Cys: Chemical Synthesis and High-Resolution X-ray Structure of ShK Toxin by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
7F4C
 
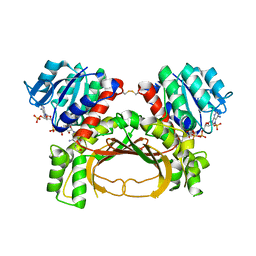 | | The crystal structure of the immature holo-enzyme of homoserine dehydrogenase complexed with NADP and 1,4-butandiol from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | 1,4-BUTANEDIOL, Homoserine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogata, K, Kaneko, R, Kubota, T, Watanabe, K, Kurihara, E, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
7F4B
 
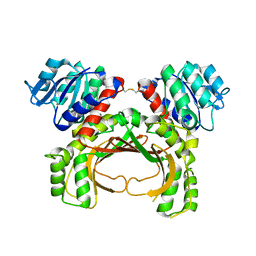 | | The crystal structure of the immature apo-enzyme of homoserine dehydrogenase from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | MAGNESIUM ION, homoserine dehydrogenase | | Authors: | Kurihara, E, Kubota, T, Watanabe, K, Ogata, K, Kaneko, R, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
5I5C
 
 | | X-ray crystal structure of allo-Thr31-ShK | | Descriptor: | GLYCEROL, Kappa-stichotoxin-She3a, LITHIUM ION, ... | | Authors: | Dang, B, Kubota, T, Manda, K.l, Shen, R, Bezanilla, F, Roux, B, Kent, S.B.H. | | Deposit date: | 2016-02-15 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inversion of the Side-Chain Stereochemistry of Indvidual Thr or Ile Residues in a Protein Molecule: Impact on the Folding, Stability, and Structure of the ShK Toxin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5I5B
 
 | | quasi racemic structure of allo-Thr13-ShK and D-ShK | | Descriptor: | D-ShK, GLYCEROL, Kappa-stichotoxin-She3a, ... | | Authors: | Dang, B, Kubota, T, Mandal, K, Shen, R, Bezanilla, F, Roux, B, Kent, S.B.H. | | Deposit date: | 2016-02-15 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Inversion of the Side-Chain Stereochemistry of Indvidual Thr or Ile Residues in a Protein Molecule: Impact on the Folding, Stability, and Structure of the ShK Toxin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5I5A
 
 | | quasi racemic structure of allo-Ile7-ShK and D-ShK | | Descriptor: | D-MALATE, D-ShK, GLYCEROL, ... | | Authors: | Dang, B, Kubota, T, Mandal, K, Shen, R, Bezanilla, F, Roux, B, Kent, S.B.H. | | Deposit date: | 2016-02-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Inversion of the Side-Chain Stereochemistry of Indvidual Thr or Ile Residues in a Protein Molecule: Impact on the Folding, Stability, and Structure of the ShK Toxin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6LFF
 
 | | transcription factor SATB1 CUTr1 domain in complex with a phosphorothioate DNA | | Descriptor: | DNA (5'-D(*GP*(C7R)P*(PST)P*AP*AP*TP*AP*TP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*(AS)P*(PST)P*(AS)P*(PST)P*TP*AP*GP*C)-3'), DNA-binding protein SATB1 | | Authors: | Akutsu, Y, Kubota, T, Yamasaki, T, Yamasaki, K. | | Deposit date: | 2019-12-02 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enhanced affinity of racemic phosphorothioate DNA with transcription factor SATB1 arising from diastereomer-specific hydrogen bonds and hydrophobic contacts.
Nucleic Acids Res., 48, 2020
|
|
7VU9
 
 | |
6A86
 
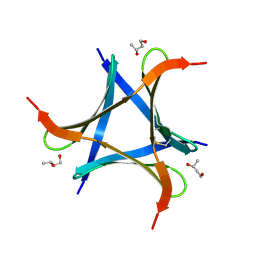 | | Pholiota squarrosa lectin | | Descriptor: | (3R)-butane-1,3-diol, lectin | | Authors: | Yamasaki, K, Yamasaki, T, Kubota, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific recognition of core fucosylation in N-glycans by Pholiota squarrosa lectin (PhoSL).
Glycobiology, 29, 2019
|
|
6A87
 
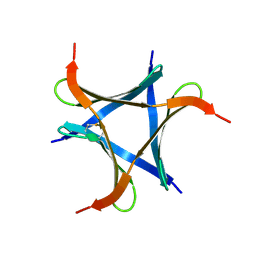 | | Pholiota squarrosa lectin (PhoSL) in complex with fucose(alpha1-6)GlcNAc | | Descriptor: | METHANETHIOL, alpha-L-fucopyranose, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamasaki, K, Yamasaki, T, Kubota, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for specific recognition of core fucosylation in N-glycans by Pholiota squarrosa lectin (PhoSL).
Glycobiology, 29, 2019
|
|
7V91
 
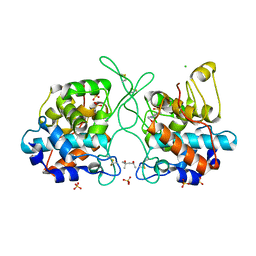 | | Crystal Structure of the Catalytic Domain of a Family GH19 Chitinase from Gazyumaru, Ficus microcarpa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kozome, D, Kubota, T, Ishikawa, K. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis and Construction of a Thermostable Antifungal Chitinase.
Appl.Environ.Microbiol., 88, 2022
|
|
7V92
 
 | |
3WBL
 
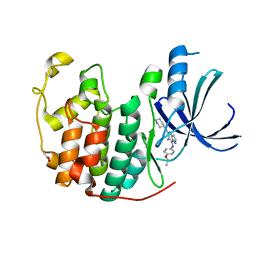 | | Crystal structure of CDK2 in complex with pyrazolopyrimidine inhibitor | | Descriptor: | ACETATE ION, Cyclin-dependent kinase 2, N~7~-(4-ethoxyphenyl)-6-methyl-N~5~-[(3S)-piperidin-3-yl]pyrazolo[1,5-a]pyrimidine-5,7-diamine | | Authors: | Fujino, A, Fukushima, K, Kubota, T, Kosugi, T, Takimoto-Kamimura, M. | | Deposit date: | 2013-05-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase-2 complex with MK2 inhibitor TEI-I01800: insight into the selectivity.
J.SYNCHROTRON RADIAT., 20, 2013
|
|
