6AB2
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oClZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-chlorophenyl)methoxycarbonylamino]hexanoic acid, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
5AVB
 
 | | human nucleosome core particle | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Wakamori, M, Fujii, Y, Umehara, T, Yokoyama, S. | | Deposit date: | 2015-06-12 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation
Sci Rep, 5, 2015
|
|
5XAX
 
 | |
5XAW
 
 | | Parallel homodimer structures of voltage-gated sodium channel beta4 for cell-cell adhesion | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Sodium channel subunit beta-4, ... | | Authors: | Shimizu, H, Yokoyama, S. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Parallel homodimer structures of the extracellular domains of the voltage-gated sodium channel beta 4 subunit explain its role in cell-cell adhesion
J. Biol. Chem., 292, 2017
|
|
1IV3
 
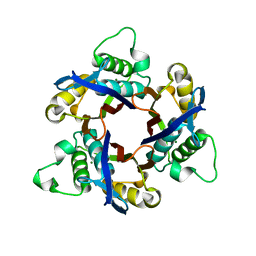 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form MG atoms) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5B6O
 
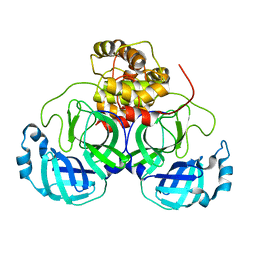 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3VQV
 
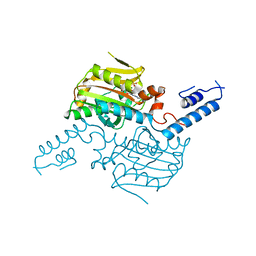 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AMPPNP (re-refined) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WQZ
 
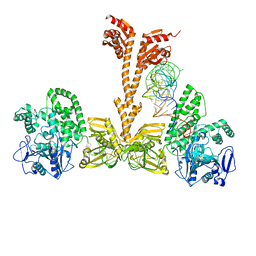 | | Crystal structure of Archaeoglobus fulgidus alanyl-tRNA synthetase in complex with a tRNA(Ala) variant having A3.U70 | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanine--tRNA ligase, RNA (75-MER), ... | | Authors: | Naganuma, M, Sekine, S, Yokoyama, S. | | Deposit date: | 2014-02-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.489 Å) | | Cite: | The selective tRNA aminoacylation mechanism based on a single G.U pair
Nature, 510, 2014
|
|
3CM0
 
 | | Crystal structure of adenylate kinase from Thermus thermophilus HB8 | | Descriptor: | Adenylate kinase | | Authors: | Nakagawa, N, Kondo, N, Masui, R, Matsuda, Z, Iwamoto, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenylate kinase from Thermus thermophilus HB8
To be Published
|
|
8I5W
 
 | | Crystal structure of the DHR-2 domain of DOCK10 in complex with Rac1 | | Descriptor: | Dedicator of cytokinesis protein 10, Ras-related C3 botulinum toxin substrate 1, SULFATE ION | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Ihara, K, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.432 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
8I5F
 
 | | Crystal structure of the DHR-2 domain of DOCK10 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10 | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
8I5V
 
 | | DOCK10 mutant L1903Y complexed with Rac1 | | Descriptor: | Dedicator of cytokinesis protein 10, Ras-related C3 botulinum toxin substrate 1, SULFATE ION | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Ihara, K, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
3WQY
 
 | | Crystal structure of Archaeoglobus fulgidus alanyl-tRNA synthetase in complex with wild-type tRNA(Ala) having G3.U70 | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Naganuma, M, Sekine, S, Yokoyama, S. | | Deposit date: | 2014-02-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The selective tRNA aminoacylation mechanism based on a single G.U pair
Nature, 510, 2014
|
|
5WRL
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y628 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WRK
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y608 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1, NICKEL (II) ION | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WRM
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y658 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
3WND
 
 | | Crystal structure of EF-Pyl | | Descriptor: | CITRIC ACID, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3WF9
 
 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate | | Descriptor: | (2S)-1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl (2S)-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3GFL
 
 | |
4NZY
 
 | | Crystal structure (type-2) of dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into Substrate Binding Mechanism in Thymidylate Kinase based on Intrinsic Dynamics of the Active Site
To be Published
|
|
3WF8
 
 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate | | Descriptor: | 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3GEZ
 
 | |
3GF2
 
 | |
2IIH
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form) | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form)
To be Published
|
|
5XJ0
 
 | | T. thermophilus RNA polymerase holoenzyme bound with gp39 and gp76 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ooi, W.Y, Murayama, Y, Mekler, V, Minakhin, L, Severinov, K, Yokoyama, S, Sekine, S. | | Deposit date: | 2017-04-28 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | A Thermus phage protein inhibits host RNA polymerase by preventing template DNA strand loading during open promoter complex formation
Nucleic Acids Res., 46, 2018
|
|
