2RRF
 
 | | The solution structure of the C-terminal region of Zinc finger FYVE domain-containing protein 21 | | Descriptor: | Zinc finger FYVE domain-containing protein 21 | | Authors: | Koshiba, S, Tomizawa, T, Hayashi, F, Tochio, N, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-08-03 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ZF21 protein, a regulator of the disassembly of focal adhesions and cancer metastasis, contains a novel noncanonical pleckstrin homology domain
J.Biol.Chem., 286, 2011
|
|
3MCJ
 
 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) other form from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3MCH
 
 | | Crystal structure of the molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, Molybdopterin biosynthesis enzyme, MoaB | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2QQ3
 
 | | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase subunit I | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426
To be Published
|
|
5H6R
 
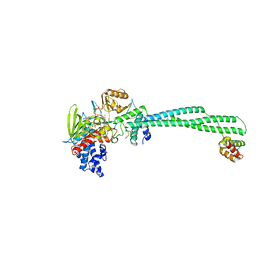 | | Crystal structure of LSD1-CoREST in complex with peptide 13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kkuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
3OPF
 
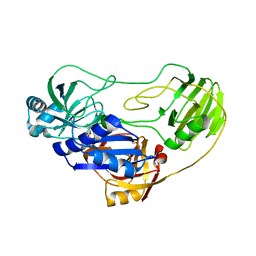 | | Crystal structure of TTHA0988 in space group P212121 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-01 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2EHU
 
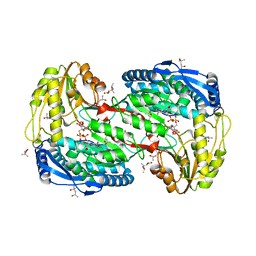 | | Crystal analysis of 1-pyrroline-5-carboxylate dehydrogenase from thermus with bound NAD and Inhibitor L-serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-09 | | Release date: | 2007-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD.
To be Published
|
|
5AX0
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.52 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AWZ
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.57 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1N27
 
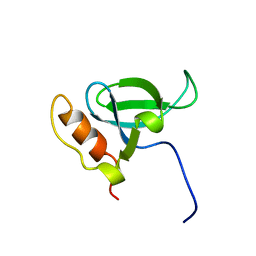 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
3ORE
 
 | | Crystal structure of TTHA0988 in space group P6522 | | Descriptor: | Putative uncharacterized protein TTHA0988 | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3VR2
 
 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR4
 
 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR6
 
 | | Crystal structure of AMP-PNP bound Enterococcus hirae V1-ATPase [bV1] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
5FWM
 
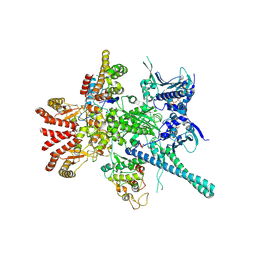 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
3VR3
 
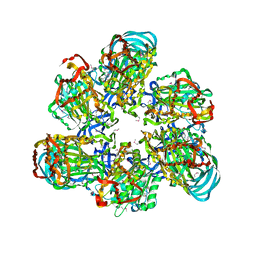 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
2RBG
 
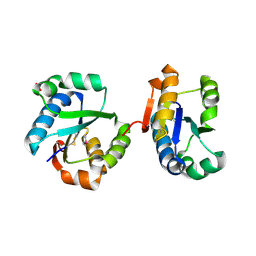 | |
2RMX
 
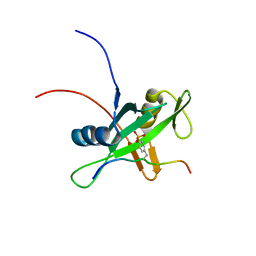 | | Solution structure of the SHP-1 C-terminal SH2 domain complexed with a tyrosine-phosphorylated peptide from NKG2A | | Descriptor: | NKG2-A/NKG2-B type II integral membrane protein, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Kasai, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of the two NKG2A immunoreceptor tyrosine-based inhibitory motifs (ITIMs) by the C-terminal SH2 domain of protein tyrosine phosphatase SHP-1
To be Published
|
|
3VR5
 
 | | Crystal structure of nucleotide-free Enterococcus hirae V1-ATPase [eV1(L)] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B, V-type sodium ATPase subunit D, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VJO
 
 | | Crystal structure of the wild-type EGFR kinase domain in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
3IEK
 
 | | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3CQ1
 
 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB138 | | Authors: | Jeyakanthan, J, Satoh, S, Kitamura, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (TT1362) from Thermus Thermophilus HB8
To be Published
|
|
3IEL
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP
To be Published
|
|
2RSM
 
 | | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65 (ICT2) | | Descriptor: | Probable peptide chain release factor C12orf65 homolog, mitochondrial | | Authors: | Enomoto, M, Tochio, N, Tomizawa, T, Koshiba, S, Guntert, P, Kigawa, T, Yokoyama, S, Nameki, N. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65.
Proteins, 2012
|
|
2ROR
 
 | | Solution structure of the VAV1 SH2 domain complexed with a tyrosine-phosphorylated peptide from SLP76 | | Descriptor: | 15-meric peptide from Lymphocyte cytosolic protein 2, Proto-oncogene vav | | Authors: | Tanaka, M, Kasai, T, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the VAV1 SH2 domain complexed with a tyrosine-phosphorylated peptide from SLP76
To be Published
|
|
