2L9V
 
 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
2LKS
 
 | | Ff11-60 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Barette, J, Velyvis, A, Religa, T.L, Korzhnev, D.M, Kay, L.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cross-Validation of the Structure of a Transiently Formed and Low Populated FF Domain Folding Intermediate Determined by Relaxation Dispersion NMR and CS-Rosetta.
J.Phys.Chem.B, 116, 2012
|
|
6WS5
 
 | | Rational drug design of phenazopyridine derivatives as novel inhibitors of Rev1-CT | | Descriptor: | 3-[(Z)-(2,3-difluorophenyl)diazenyl]pyridine-2,6-diamine, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | McPherson, K.S, Korzhnev, D.M. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structure-Based Drug Design of Phenazopyridine Derivatives as Inhibitors of Rev1 Interactions in Translesion Synthesis.
Chemmedchem, 16, 2021
|
|
6WS0
 
 | |
6BCD
 
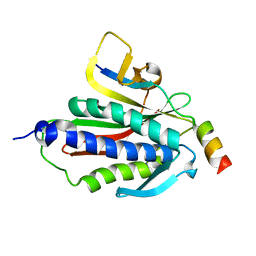 | | Crystal structure of Rev7-K44A/R124A/A135D in complex with Rev3-RBM2 (residues 1988-2014) | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BI7
 
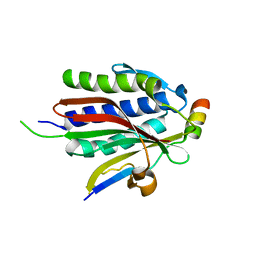 | | Crystal structure of Rev7-WT/Rev3 as a monomer under high-salt conditions | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Rizzo, A.A, Korzhnev, D.M, Hao, B, Li, Y. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BC8
 
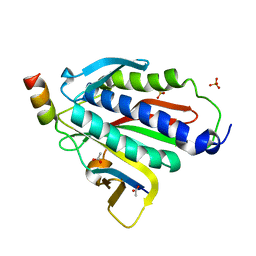 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | Descriptor: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2M85
 
 | | PHD Domain from Human SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, ZINC ION | | Authors: | Machado, L.E.S.F, Pustovalova, Y, Pozhidaeva, A, Almeida, F.C.L, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2013-05-07 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PHD domain from human SHPRH.
J.Biomol.Nmr, 56, 2013
|
|
1RQT
 
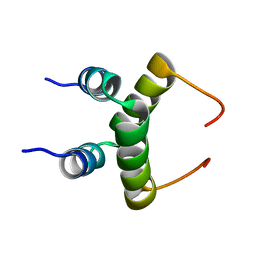 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
2M2I
 
 | |
1RQU
 
 | | NMR structure of L7 dimer from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
2MRE
 
 | | NMR structure of the Rad18-UBZ/ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Polyubiquitin-C, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2MRF
 
 | | NMR structure of the ubiquitin-binding zinc finger (UBZ) domain from human Rad18 | | Descriptor: | E3 ubiquitin-protein ligase RAD18, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
1RQV
 
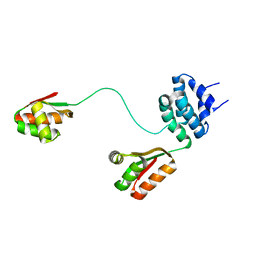 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
1RQS
 
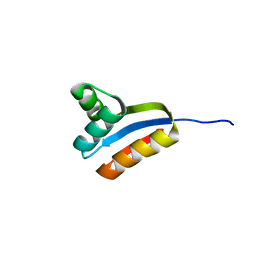 | | NMR structure of C-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
