3ND2
 
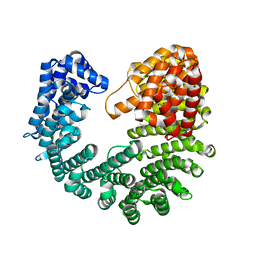 | | Structure of Yeast Importin-beta (Kap95p) | | Descriptor: | Importin subunit beta-1 | | Authors: | Forwood, J.K, Kobe, B. | | Deposit date: | 2010-06-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Quantitative Structural Analysis of Importin-beta Flexibility: Paradigm for Solenoid Protein Structures
Structure, 18, 2010
|
|
2V1O
 
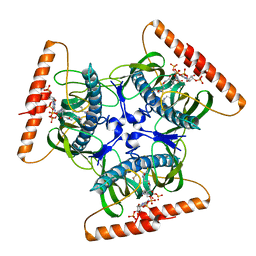 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5I34
 
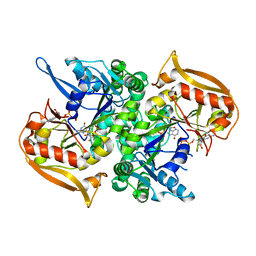 | | Adenylosuccinate synthetase from Cryptococcus neoformans complexed with GDP and IMP | | Descriptor: | Adenylosuccinate synthetase, GUANOSINE-5'-DIPHOSPHATE, INOSINIC ACID | | Authors: | Blundell, R.D, Williams, S.J, Ericsson, D, Fraser, J.A, Kobe, B. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Disruption of de Novo Adenosine Triphosphate (ATP) Biosynthesis Abolishes Virulence in Cryptococcus neoformans.
Acs Infect Dis., 2, 2016
|
|
3L3Q
 
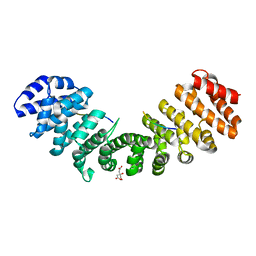 | | Mouse importin alpha-pepTM NLS peptide complex | | Descriptor: | CITRATE ANION, Importin subunit alpha-2, pepTM | | Authors: | Takeda, A.A.S, Kobe, B, Fontes, M.R.M. | | Deposit date: | 2009-12-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening.
J.Biol.Chem., 285, 2010
|
|
6CSL
 
 | | Pneumococcal PhtD protein 269-339 fragment with bound Zn(II) | | Descriptor: | Histidine triad protein D, ZINC ION | | Authors: | Luo, Z, Pederick, V.G, Paton, J.C, McDevitt, C.A, Kobe, B. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural characterisation of the HT3 motif of the polyhistidine triad protein D from Streptococcus pneumoniae.
FEBS Lett., 592, 2018
|
|
7BER
 
 | | SFX structure of the MyD88 TIR domain higher-order assembly (solved, rebuilt and refined using an identical protocol to the MicroED structure of the MyD88 TIR domain higher-order assembly) | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7BEQ
 
 | | MicroED structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
1EJL
 
 | |
5I33
 
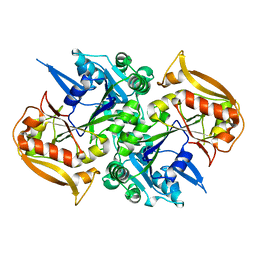 | | Unligated adenylosuccinate synthetase from Cryptococcus neoformans | | Descriptor: | Adenylosuccinate synthetase | | Authors: | Blundell, R.D, Williams, S.J, Ericsson, D, Fraser, J.A, Kobe, B. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of de Novo Adenosine Triphosphate (ATP) Biosynthesis Abolishes Virulence in Cryptococcus neoformans.
Acs Infect Dis., 2, 2016
|
|
3ZLH
 
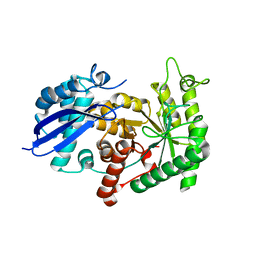 | | Structure of group A Streptococcal enolase | | Descriptor: | ENOLASE | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
1WNH
 
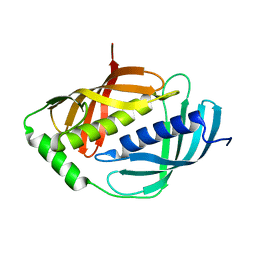 | | Crystal structure of mouse Latexin (tissue carboxypeptidase inhibitor) | | Descriptor: | Latexin | | Authors: | Aagaard, A, Listwan, P, Cowieson, N, Huber, T, Ravasi, T, Wells, C.A, Flanagan, J.U, Hume, D.A, Kobe, B, Martin, J.L. | | Deposit date: | 2004-08-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Inflammatory Role for the Mammalian Carboxypeptidase Inhibitor Latexin: Relationship to Cystatins and the Tumor Suppressor TIG1
Structure, 13, 2005
|
|
3ZTT
 
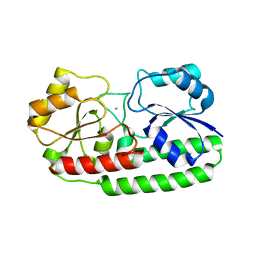 | | Crystal structure of pneumococcal surface antigen PsaA with manganese | | Descriptor: | MANGANESE (II) ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | McDevitt, C.A, Ogunniyi, A.D, Valkov, E, Lawrence, M.C, Kobe, B, McEwan, A.G, Paton, J.C. | | Deposit date: | 2011-07-12 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular mechanism for bacterial susceptibility to zinc.
PLoS Pathog., 7, 2011
|
|
4OIH
 
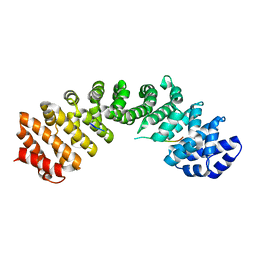 | | Importin Alpha in Complex with the Bipartite NLS of Prp20 | | Descriptor: | Guanine nucleotide exchange factor SRM1, Importin subunit alpha-1 | | Authors: | Roman, N, Christie, M, Swarbrick, C.M.D, Kobe, B, Forwood, J.K. | | Deposit date: | 2014-01-19 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterisation of the Nuclear Import Receptor Importin Alpha in Complex with the Bipartite NLS of Prp20
Plos One, 8, 2013
|
|
1PJN
 
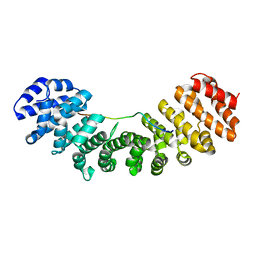 | | Mouse Importin alpha-bipartite NLS N1N2 from Xenopus laevis phosphoprotein Complex | | Descriptor: | Histone-binding protein N1/N2, Importin alpha-2 subunit | | Authors: | Fontes, M.R.M, Teh, T, Jans, D, Brinkworth, R.I, Kobe, B. | | Deposit date: | 2003-06-03 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha
J.Biol.Chem., 278, 2003
|
|
1PJM
 
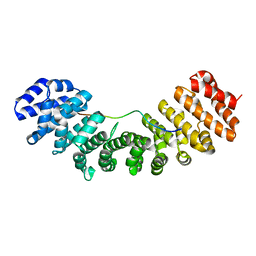 | | Mouse Importin alpha-bipartite NLS from human retinoblastoma protein Complex | | Descriptor: | Importin alpha-2 subunit, Retinoblastoma-associated protein | | Authors: | Fontes, M.R.M, Teh, T, Jans, D, Brinkworth, R.I, Kobe, B. | | Deposit date: | 2003-06-03 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha
J.Biol.Chem., 278, 2003
|
|
6WES
 
 | | Crystal structure of the effector SnTox3 from Parastagonospora nodorum | | Descriptor: | Tox3 | | Authors: | Outram, M.A, Williams, S.J, Ericsson, D.J, Kobe, B, Solomon, P.S. | | Deposit date: | 2020-04-02 | | Release date: | 2020-11-04 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The crystal structure of SnTox3 from the necrotrophic fungus Parastagonospora nodorum reveals a unique effector fold and provides insight into Snn3 recognition and pro-domain protease processing of fungal effectors.
New Phytol., 231, 2021
|
|
7UWG
 
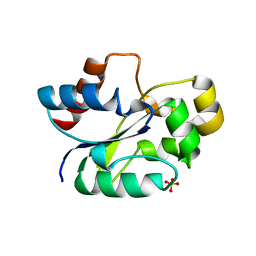 | | The crystal structure of the TIR domain-containing protein from Acinetobacter baumannii (AbTir) | | Descriptor: | HEXAETHYLENE GLYCOL, Molecular chaperone Tir, SULFATE ION | | Authors: | Manik, M.K, Nanson, J.D, Ve, T, Kobe, B. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXU
 
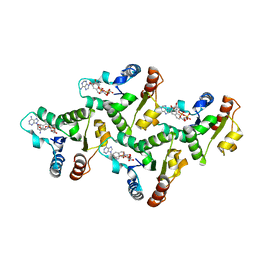 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | Descriptor: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
2OPC
 
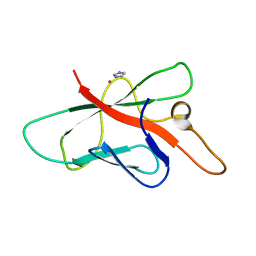 | | Structure of Melampsora lini avirulence protein, AvrL567-A | | Descriptor: | AvrL567-A, COBALT (II) ION, IMIDAZOLE | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-01-29 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3ZLG
 
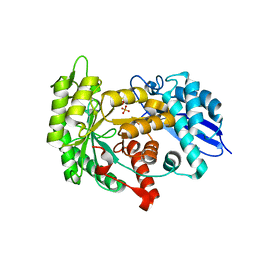 | | Structure of group A Streptococcal enolase K362A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
3OZI
 
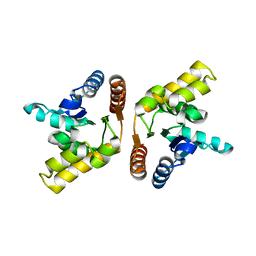 | | Crystal structure of the TIR domain from the flax disease resistance protein L6 | | Descriptor: | COBALT (II) ION, L6tr | | Authors: | Ve, T, Bernoux, M, Williams, S, Valkov, E, Warren, C, Hatters, D, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2010-09-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation.
Cell Host Microbe, 9, 2011
|
|
6VZB
 
 | | Crystal structure of cytochrome P450 NasF5053 S284A-V288A mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VXV
 
 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
7SZL
 
 | | Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1R10 (IL-1RAPL2) | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Nimma, S, Gu, W, Manik, M.K, Ve, T, Nanson, J.D, Kobe, B. | | Deposit date: | 2021-11-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Toll/interleukin-1 receptor (TIR) domain of IL-1R10 provides structural insights into TIR domain signalling.
Febs Lett., 596, 2022
|
|
4FH0
 
 | | Crystal Structure of Human BinCARD CARD, double mutant F16M/L66M SeMet form | | Descriptor: | Bcl10-interacting CARD protein, SULFATE ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2012-06-05 | | Release date: | 2013-02-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
