2MH9
 
 | |
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
8KE8
 
 | | Crystal structure of TetR-type transcriptional factor NalC from P. aeruginosa | | Descriptor: | NalC | | Authors: | Lee, J.Y, Jeong, K.H, Ko, J.H, Son, S.B. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the transcriptional regulator NalC, a key component of the MexAB-OprM efflux pump system, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 679, 2023
|
|
9J8E
 
 | | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor | | Descriptor: | BIOTINYL-5-AMP, Bifunctional ligase/repressor BirA | | Authors: | Lee, J.Y, Jeong, K.H, Son, S.B, Ko, J.H. | | Deposit date: | 2024-08-21 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9J8F
 
 | | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Bifunctional ligase/repressor BirA, PENTAETHYLENE GLYCOL | | Authors: | Lee, J.Y, Jeong, K.H, Son, S.B, Ko, J.H. | | Deposit date: | 2024-08-21 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
6TL8
 
 | | Structural basis of SALM3 dimerization and adhesion complex formation with the presynaptic receptor protein tyrosine phosphatases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Myeloid cell surface antigen CD33,Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | | Authors: | Karki, S, Shkumatov, A.V, Bae, S, Ko, J, Kajander, T. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of SALM3 dimerization and synaptic adhesion complex formation with PTP sigma.
Sci Rep, 10, 2020
|
|
4XTT
 
 | | Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP) | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Putative potassium transport protein | | Authors: | Kim, H, Youn, S.J, Kim, S.O, Ko, J, Lee, J.O, Choi, B.S. | | Deposit date: | 2015-01-24 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C Domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP)
J.Biol.Chem., 290, 2015
|
|
3KYG
 
 | | Crystal structure of VCA0042 (L135R) complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein VCA0042 | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
3KYF
 
 | | Crystal structure of P4397 complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
4LIS
 
 | | Crystal Structure of UDP-galactose-4-epimerase from Aspergillus nidulans | | Descriptor: | GLYCEROL, IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Ko, J, Sheoran, I, Kaminskyj, S.G.W, Sanders, D.A.R. | | Deposit date: | 2013-07-03 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidation of Substrate Specificity in Aspergillus nidulans UDP-Galactose-4-Epimerase.
Plos One, 8, 2013
|
|
2LSI
 
 | |
4EXT
 
 | |
5AXH
 
 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose | | Descriptor: | Dextranase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
5AXG
 
 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Dextranase, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
6RLE
 
 | | Crystal structure of human monoamine oxidase B in complex with styrylpiperidine analogue 97 | | Descriptor: | 4-[2-(4-propan-2-ylphenyl)ethyl]-1-[(~{E})-prop-1-enyl]piperidine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Knez, D, Colettis, N, Sova, M, Pislar, A, Higgs, J, Kamecki, F, Mangialavori, I, Dolsak, A, Zakelj, S, Trontelj, J, Kos, J, Marder, N.M, Gobec, S, Binda, C. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B.
J.Med.Chem., 63, 2020
|
|
6RKB
 
 | | Crystal structure of human monoamine oxidase B in complex with styrylpiperidine analogue 1 | | Descriptor: | 4-[(~{E})-2-(4-fluorophenyl)ethenyl]-1-[(~{E})-prop-1-enyl]piperidine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Knez, D, Colettis, N, Sova, M, Pislar, A, Higgs, J, Kamecki, F, Mangialavori, I, Dolsak, A, Zakelj, S, Trontelj, J, Kos, J, Marder, N.M, Gobec, S, Binda, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B.
J.Med.Chem., 63, 2020
|
|
6RKP
 
 | | Crystal structure of human monoamine oxidase B in complex with styrylpiperidine analogue 84 | | Descriptor: | 1-[(~{E})-prop-1-enyl]-4-[(~{E})-2-[4-(trifluoromethyl)phenyl]ethenyl]piperidine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Knez, D, Colettis, N, Sova, M, Pislar, A, Higgs, J, Kamecki, F, Mangialavori, I, Dolsak, A, Zakelj, S, Trontelj, J, Kos, J, Marder, N.M, Gobec, S, Binda, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B.
J.Med.Chem., 63, 2020
|
|
3NBE
 
 | | Clitocybe nebularis ricin B-like lectin (CNL) in complex with N,N'-diacetyllactosediamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-2-azidoethyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Ricin B-like lectin, SULFATE ION | | Authors: | Renko, M, Pohleven, J, Sabotic, J, Kos, J, Turk, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,N'-diacetyllactosediamine (GalNAc beta 1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis
J.Biol.Chem., 287, 2012
|
|
3N0K
 
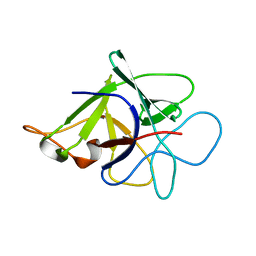 | | Proteinase inhibitor from Coprinopsis cinerea | | Descriptor: | Serine protease inhibitor 1 | | Authors: | Renko, M, Sabotic, J, Bleuler-Martinez, S, Kallert, S, Avanzo, P, Kos, J, Aebi, M, Kuenzler, M, Turk, D. | | Deposit date: | 2010-05-14 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Trypsin Inhibition and Entomotoxicity of Cospin, Serine Protease Inhibitor Involved in Defense of Coprinopsis cinerea Fruiting Bodies.
J.Biol.Chem., 287, 2012
|
|
3NBD
 
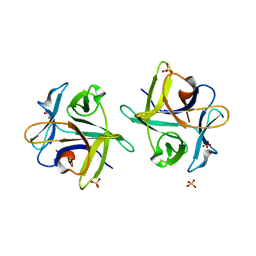 | | Clitocybe nebularis ricin B-like lectin (CNL) in complex with lactose, crystallized at pH 7.1 | | Descriptor: | Ricin B-like lectin, SULFATE ION, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Renko, M, Pohleven, J, Sabotic, J, Kos, J, Turk, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,N'-diacetyllactosediamine (GalNAc beta 1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis
J.Biol.Chem., 287, 2012
|
|
3NBC
 
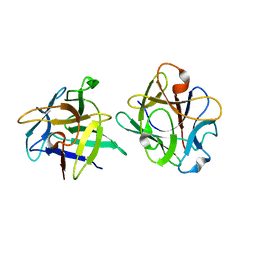 | | Clitocybe nebularis ricin B-like lectin (CNL) in complex with lactose, crystallized at pH 4.4 | | Descriptor: | Ricin B-like lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Renko, M, Pohleven, J, Sabotic, J, Kos, J, Turk, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,N'-diacetyllactosediamine (GalNAc beta 1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis
J.Biol.Chem., 287, 2012
|
|
1A90
 
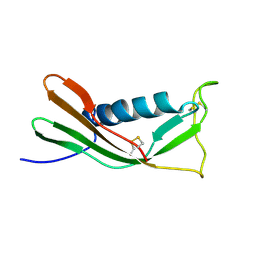 | | RECOMBINANT MUTANT CHICKEN EGG WHITE CYSTATIN, NMR, 31 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-04-14 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
1A67
 
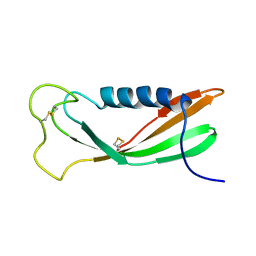 | | CHICKEN EGG WHITE CYSTATIN WILDTYPE, NMR, 16 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-03-06 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
4ION
 
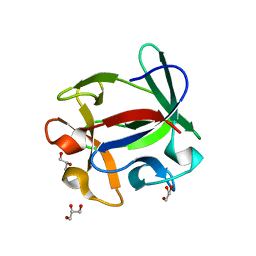 | | Macrolepiota procera ricin B-like lectin (MPL) | | Descriptor: | GLYCEROL, Ricin B-like lectin | | Authors: | Renko, M, Zurga, S, Sabotic, J, Pohleven, J, Kos, J, Turk, D. | | Deposit date: | 2013-01-08 | | Release date: | 2013-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Macrolepiota procera ricin B-like lectin (MPL)
TO BE PUBLISHED
|
|
