3T8C
 
 | |
3RM0
 
 | | Human Thrombin in complex with MI354 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RMM
 
 | | Human Thrombin in complex with MI332 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3T7P
 
 | | Endothiapepsin in complex with a hydrazide derivative | | Descriptor: | (2R)-2-amino-N'-[(E)-(4-hydroxynaphthalen-1-yl)methylidene]-2-phenylethanehydrazide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2011-07-30 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Endothiapepsin in complex with a hydrazide derivative
To be Published
|
|
3T74
 
 | | Thermolysin In Complex With UBTLN27 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Water makes the difference: rearrangement of water solvation layer triggers non-additivity of functional group contributions in protein-ligand binding.
Chemmedchem, 7, 2012
|
|
3T6I
 
 | | Endothiapepsin in complex with an azepin derivative | | Descriptor: | (3R)-3-({(4-aminobenzyl)[(4-aminophenyl)acetyl]amino}methyl)-5-(hydroxymethyl)-2,3,4,7-tetrahydro-1H-azepinium, (3R)-3-({(4-aminobenzyl)[(4-aminophenyl)acetyl]amino}methyl)-5-{[(4-bromobenzoyl)oxy]methyl}-2,3,4,7-tetrahydro-1H-azepinium, 4-bromobenzoic acid, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Endothiapepsin in complex with an azepin derivative
To be Published
|
|
3T8F
 
 | | Thermolysin In Complex With UBTLN34 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-08-01 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Water makes the difference: rearrangement of water solvation layer triggers non-additivity of functional group contributions in protein-ligand binding.
Chemmedchem, 7, 2012
|
|
3RMO
 
 | | Human Thrombin in complex with MI004 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RML
 
 | | Human Thrombin in complex with MI331 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3SHC
 
 | | Human Thrombin In Complex With UBTHR101 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(4-chloropyridin-2-yl)methyl]-L-prolinamide, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-06-16 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
3SI3
 
 | | Human Thrombin In Complex With UBTHR103 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-(pyridin-2-ylmethyl)-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
3U98
 
 | | Human Thrombin In Complex With MI001 | | Descriptor: | (2S)-1-[(2R)-2-(benzylsulfonylamino)-5-guanidino-pentanoyl]-N-[(4-carbamimidoylphenyl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-10-18 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
3U9A
 
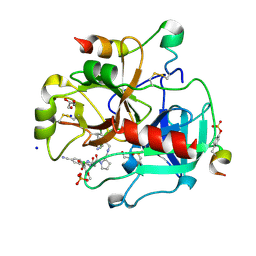 | | Human Thrombin In Complex With MI330 | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloranyl-phenyl]methyl]-1-[(2S)-2-[(3-chloranyl-4-methoxy-phenyl)sulfonylamino]-4-[(4-cyanophenyl)methylamino]-4-oxidanylidene-butanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-10-18 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
3LJO
 
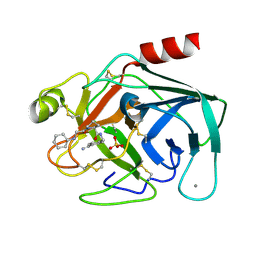 | | Bovine trypsin in complex with UB-THR 11 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Wegscheid-Gerlach, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties.
J.Mol.Biol., 405, 2011
|
|
3LBO
 
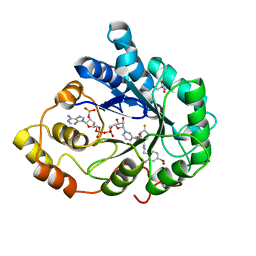 | | Human aldose reductase mutant T113C complexed with IDD594 | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-08 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LEN
 
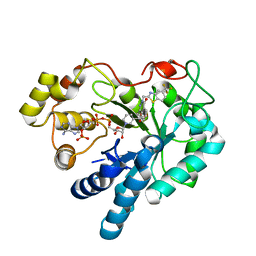 | | Human Aldose Reductase mutant T113S complexed with Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-15 | | Release date: | 2011-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
3LQL
 
 | | Human Aldose Reductase mutant T113A complexed with IDD 594 | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-02-09 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LD5
 
 | | Human aldose reductase mutant T113S complexed with IDD594 | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LZ3
 
 | | Human aldose reductase mutant T113S complexed with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-01 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LZ5
 
 | | Human aldose reductase mutant T113V complexed with IDD594 | | Descriptor: | Aldose Reductase, CITRIC ACID, IDD594, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-01 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3M14
 
 | | Carbonic Anhydrase II in complex with novel sulfonamide inhibitor | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, N-[(2Z)-1,3-thiazolidin-2-ylidene]sulfamide, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G, Sandner, N.U. | | Deposit date: | 2010-03-04 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural investigation and inhibitor studies on Carbonic Anhydrase II
To be Published
|
|
3LZY
 
 | |
3MB9
 
 | | Human Aldose Reductase mutant T113A complexed with Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-25 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
3MC5
 
 | |
3M1W
 
 | |
