5UQG
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p200 | | 分子名称: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, INOSINIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-08 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p200
To Be Published
|
|
5V4D
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis | | 分子名称: | ACETIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-03-09 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis
To Be Published
|
|
5URS
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P178 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-12 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.388 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P178
To Be Published
|
|
5UUV
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with a product IMP and the inhibitor P182 | | 分子名称: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-17 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with a product IMP and the inhibitor P182
To Be Published
|
|
5V4F
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis | | 分子名称: | GLYCEROL, Putative translational inhibitor protein | | 著者 | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-03-09 | | 公開日 | 2017-04-05 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis
To Be Published
|
|
5UQH
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p182 | | 分子名称: | 1,2-ETHANEDIOL, INOSINIC ACID, ISOPROPYL ALCOHOL, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-08 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
7F27
 
 | | Crystal structure of polyketide ketosynthase | | 分子名称: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | 著者 | Kim, Y, Lee, W.C. | | 登録日 | 2021-06-10 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.806 Å) | | 主引用文献 | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
5UQF
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with IMP and the inhibitor P225 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-08 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Campylobacter jejuni in the complex with IMP and the inhibitor P225
To Be Published
|
|
5UUZ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P200 | | 分子名称: | 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-17 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.496 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P200
To Be Published
|
|
5URQ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-alpha-D-ribofuranosylamine, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-12 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176
To Be Published
|
|
5UME
 
 | | Crystal Structure of 5,10-Methylenetetrahydrofolate Reductase MetF from Haemophilus influenzae | | 分子名称: | 1,2-ETHANEDIOL, 5,10-methylenetetrahydrofolate reductase, ACETIC ACID, ... | | 著者 | Kim, Y, Mulligan, R, Maltseva, N, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-01-27 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of 5,10-Methylenetetrahydrofolate Reductase MetF from Haemophilus influenzae
To Be Published
|
|
5UPU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | 著者 | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Mulligan, R, Gu, M, Sacchettini, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-04 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.905 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
5UPY
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21 | | 分子名称: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-04 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21
To Be Published
|
|
5UPV
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36 | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-04 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis In the presence of G36
To Be Published
|
|
5UVE
 
 | | Crystal Structure of the ABC Transporter Substrate-binding protein BAB1_0226 from Brucella abortus | | 分子名称: | CALCIUM ION, GLYCEROL, Substrate-binding region of ABC-type glycine betaine transport system | | 著者 | Kim, Y, Chhor, G, Endres, M, Hero, J, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-02-20 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Beta-barrel-like Protein of Unknown Function
To Be Published
|
|
5UPX
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the presence of Xanthosine Monophosphate | | 分子名称: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, XANTHOSINE-5'-MONOPHOSPHATE | | 著者 | Kim, Y, Makowska-Grzyska, M, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-04 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.855 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the presence of Xanthosine Monophosphate
To Be Published
|
|
5WHM
 
 | | Crystal Structure of IclR Family Transcriptional Regulator from Brucella abortus | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | 著者 | Kim, Y, Wu, R, Tesar, C, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-07-17 | | 公開日 | 2017-08-23 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Molecular control of gene expression byBrucellaBaaR, an IclR-type transcriptional repressor.
J. Biol. Chem., 293, 2018
|
|
5VVH
 
 | | Crystal Structure of the Effector Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens | | 分子名称: | FORMIC ACID, Octopine catabolism/uptake operon regulatory protein OccR, SULFATE ION | | 著者 | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-05-19 | | 公開日 | 2017-06-21 | | 最終更新日 | 2018-09-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
1YB4
 
 | | Crystal Structure of the Tartronic Semialdehyde Reductase from Salmonella typhimurium LT2 | | 分子名称: | tartronic semialdehyde reductase | | 著者 | Kim, Y, Wu, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-12-20 | | 公開日 | 2005-02-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
1AT9
 
 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | 分子名称: | BACTERIORHODOPSIN, RETINAL | | 著者 | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | 登録日 | 1997-08-20 | | 公開日 | 1998-09-16 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | 主引用文献 | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
2XMS
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | 分子名称: | CHLORIDE ION, IMIDAZOLE, PROTEIN NDRG2 | | 著者 | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | 登録日 | 2010-07-29 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2XMR
 
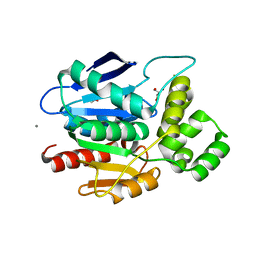 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | 登録日 | 2010-07-29 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
5VSV
 
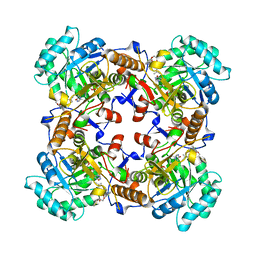 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P225 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, {2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenoxy}acetic acid | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-05-12 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.205 Å) | | 主引用文献 | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P225
To Be Published
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | 分子名称: | 3C-like proteinase, CHLORIDE ION | | 著者 | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | 登録日 | 2021-10-08 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation
To Be Published
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | 登録日 | 2021-10-13 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation
To Be Published
|
|
