4LW2
 
 | |
2QDF
 
 | | Structure of N-terminal domain of E. Coli YaeT | | Descriptor: | MAGNESIUM ION, Outer membrane protein assembly factor yaeT | | Authors: | Kim, S, Malinverni, J.C, Sliz, P, Silhavy, T.J, Harrison, S.C, Kahne, D. | | Deposit date: | 2007-06-20 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of an essential component of the outer membrane protein assembly machine.
Science, 317, 2007
|
|
2QCZ
 
 | | Structure of N-terminal domain of E. Coli YaeT | | Descriptor: | Outer membrane protein assembly factor yaeT | | Authors: | Kim, S, Malinverni, J.C, Sliz, P, Silhavy, T.J, Harrison, S.C, Kahne, D. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of an essential component of the outer membrane protein assembly machine.
Science, 317, 2007
|
|
3ACP
 
 | | Crystal Structure of Yeast Rpn14, a Chaperone of the 19S Regulatory Particle of the Proteasome | | Descriptor: | WD repeat-containing protein YGL004C | | Authors: | Kim, S, Saeki, Y, Suzuki, A, Takagi, K, Fukunaga, K, Yamane, T, Kato, K, Tanaka, K, Mizushima, T. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast Rpn14, a chaperone of the 19S regulatory particle of the proteasome
J.Biol.Chem., 285, 2010
|
|
7UIY
 
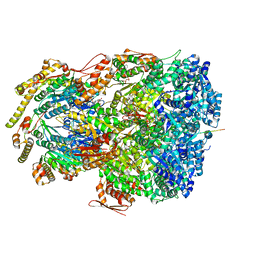 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
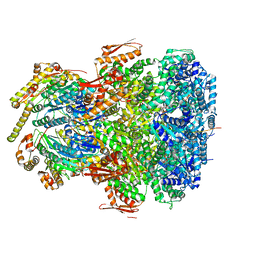 | | ClpAP complex bound to ClpS N-terminal extension, class I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
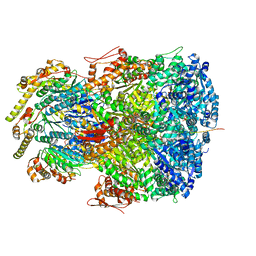 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
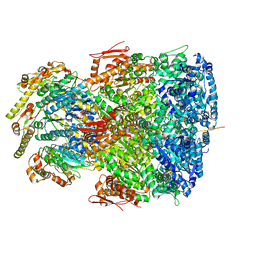 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
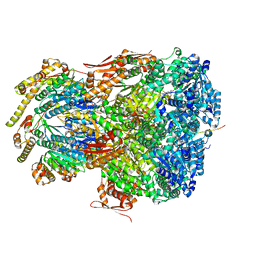 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
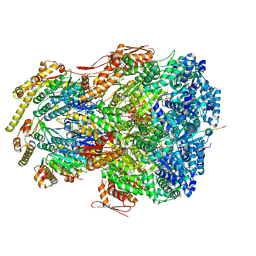 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VID
 
 | |
3VL1
 
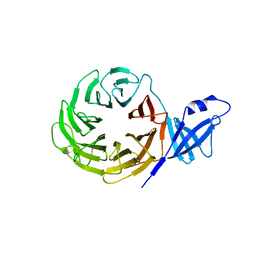 | | Crystal structure of yeast Rpn14 | | Descriptor: | 26S proteasome regulatory subunit RPN14 | | Authors: | Kim, S, Nishide, A, Saeki, Y, Takagi, K, Tanaka, K, Kato, K, Mizushima, T. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3OTP
 
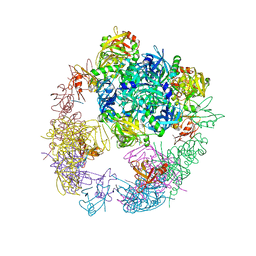 | |
2MLP
 
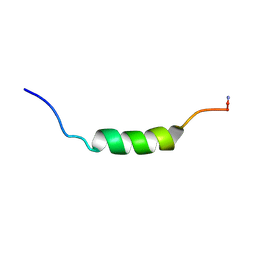 | | MICROCIN LEADER PEPTIDE FROM E. COLI, NMR, 25 STRUCTURES | | Descriptor: | MCBA PROPEPTIDE | | Authors: | Kim, S, Sinha Roy, R, Walsh, C.T, Baleja, J.D. | | Deposit date: | 1998-01-21 | | Release date: | 1998-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26.
Chem.Biol., 5, 1998
|
|
1SJ0
 
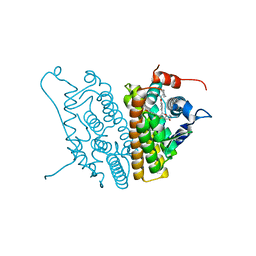 | | Human Estrogen Receptor Alpha Ligand-binding Domain in Complex with the Antagonist Ligand 4-D | | Descriptor: | (2S,3R)-2-(4-(2-(PIPERIDIN-1-YL)ETHOXY)PHENYL)-2,3-DIHYDRO-3-(4-HYDROXYPHENYL)BENZO[B][1,4]OXATHIIN-6-OL, Estrogen receptor | | Authors: | Kim, S, Wu, J.Y, Birzin, E.T, Chan, W, Pai, L.Y, Yang, Y.T, Mosley, R.T, Fitzgerald, P.M, Sharma, N, DiNinno, F, Rohrer, S.P, Schaeffer, J.M, Hammond, M.L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor alpha Modulators.
J.Med.Chem., 47, 2004
|
|
1R1A
 
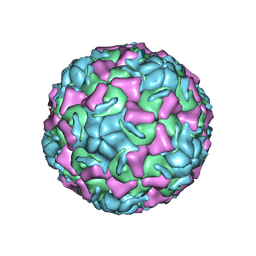 | | CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS SEROTYPE 1A (HRV1A) | | Descriptor: | HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP2), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Kim, S, Rossmann, M.G. | | Deposit date: | 1989-03-15 | | Release date: | 1990-07-15 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human rhinovirus serotype 1A (HRV1A).
J.Mol.Biol., 210, 1989
|
|
1EUJ
 
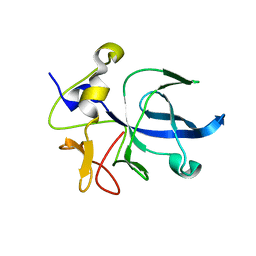 | | A NOVEL ANTI-TUMOR CYTOKINE CONTAINS A RNA-BINDING MOTIF PRESENT IN AMINOACYL-TRNA SYNTHETASES | | Descriptor: | ENDOTHELIAL MONOCYTE ACTIVATING POLYPEPTIDE 2 | | Authors: | Kim, Y, Shin, J, Li, R, Cheong, C, Kim, S. | | Deposit date: | 2000-04-17 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel anti-tumor cytokine contains an RNA binding motif present in aminoacyl-tRNA synthetases.
J.Biol.Chem., 275, 2000
|
|
5JJH
 
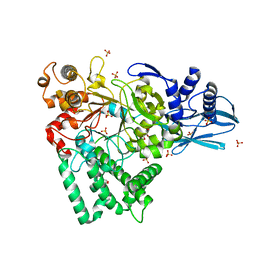 | |
6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
6JQE
 
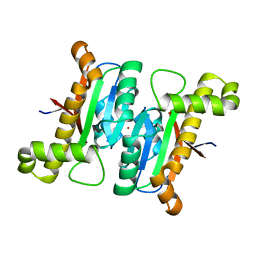 | | The structural basis of the beta-carbonic anhydrases CafD (wild type) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
6JQC
 
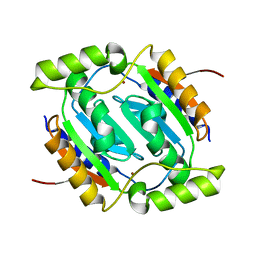 | | The structural basis of the beta-carbonic anhydrase CafC (wild type) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
6JQD
 
 | | The structural basis of the beta-carbonic anhydrase CafC (L25G and L78G mutant) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
8OSB
 
 | | TWIST1-TCF4-ALX4 complex on specific DNA | | Descriptor: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | Authors: | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
7C11
 
 | | Formate--tetrahydrofolate ligase from Methylobacterium extorquens CM4 strain | | Descriptor: | ACETATE ION, CITRATE ANION, Formate-tetrahydrofolate ligase, ... | | Authors: | Kim, K.-J, Kim, S, Seo, H, Lee, S. | | Deposit date: | 2020-05-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.815 Å) | | Cite: | Biochemical properties and crystal structure of formate-tetrahydrofolate ligase from Methylobacterium extorquens CM4.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
1FYJ
 
 | | SOLUTION STRUCTURE OF MULTI-FUNCTIONAL PEPTIDE MOTIF-1 PRESENT IN HUMAN GLUTAMYL-PROLYL TRNA SYNTHETASE (EPRS). | | Descriptor: | MULTIFUNCTIONAL AMINOACYL-TRNA SYNTHETASE | | Authors: | Jeong, E.-J, Hwang, G.-S, Kim, K.H, Kim, M.J, Kim, S, Kim, K.-S. | | Deposit date: | 2000-10-02 | | Release date: | 2001-03-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of multifunctional peptide motifs in human bifunctional tRNA synthetase: identification of RNA-binding residues and functional implications for tandem repeats.
Biochemistry, 39, 2000
|
|
