1L7P
 
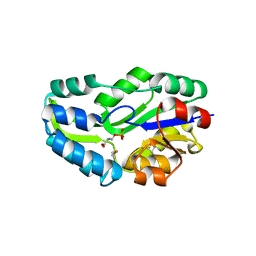 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
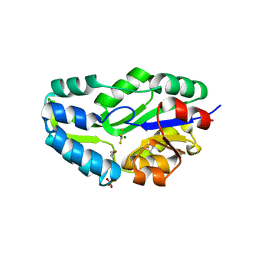 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L2F
 
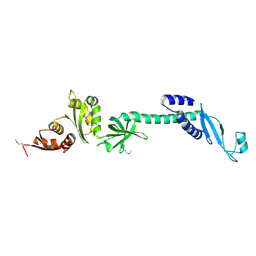 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
1LFP
 
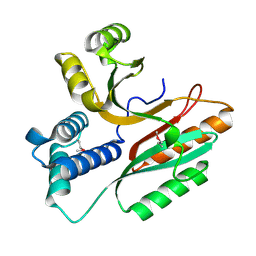 | | Crystal Structure of a Conserved Hypothetical Protein Aq1575 from Aquifex Aeolicus | | Descriptor: | Hypothetical protein AQ_1575 | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L7O
 
 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
2A4L
 
 | |
2ZC8
 
 | | Crystal structure of N-Acylamino Acid Racemase from Thermus thermophilus HB8 | | Descriptor: | N-acylamino acid racemase | | Authors: | Hayashida, M, Kim, S.H, Takeda, K, Hisano, T, Miki, K. | | Deposit date: | 2007-11-05 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-acylamino acid racemase from Thermus thermophilus HB8
Proteins, 71, 2008
|
|
1PF8
 
 | | Crystal Structure of Human Cyclin-Dependent Kinase 2 Complexed with a Nucleoside Inhibitor | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, Cell division protein kinase 2 | | Authors: | Moshinsky, D.J, Bellamacina, R.C, Boisvert, D.C, Huang, P, Hui, T, Jancarik, J, Kim, S.H, Rice, A.G. | | Deposit date: | 2003-05-24 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | SU9516: biochemical analysis of cdk inhibition and crystal structure in complex with cdk2.
Biochem.Biophys.Res.Commun., 310, 2003
|
|
1LXD
 
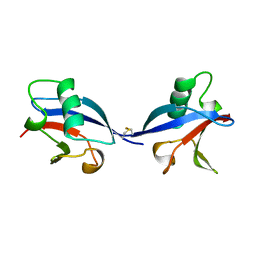 | | CRYSTAL STRUCTURE OF THE RAS INTERACTING DOMAIN OF RALGDS, A GUANINE NUCLEOTIDE DISSOCIATION STIMULATOR OF RAL PROTEIN | | Descriptor: | RALGDSB | | Authors: | Huang, L, Weng, X.W, Hofer, F, Martin, G.S, Kim, S.H. | | Deposit date: | 1997-03-05 | | Release date: | 1998-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the Ras-interacting domain of RalGDS.
Nat.Struct.Biol., 4, 1997
|
|
1JOW
 
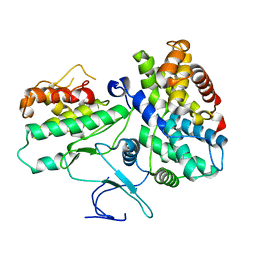 | |
3R4Y
 
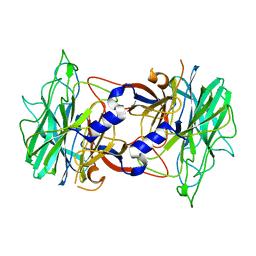 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
1B78
 
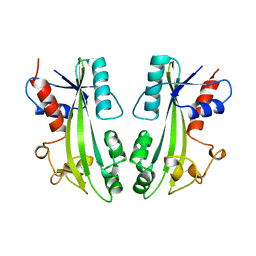 | | STRUCTURE-BASED IDENTIFICATION OF THE BIOCHEMICAL FUNCTION OF A HYPOTHETICAL PROTEIN FROM METHANOCOCCUS JANNASCHII:MJ0226 | | Descriptor: | PYROPHOSPHATASE | | Authors: | Hwang, K.Y, Chung, J.H, Han, Y.S, Kim, S.H, Cho, Y. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based identification of a novel NTPase from Methanococcus jannaschii.
Nat.Struct.Biol., 6, 1999
|
|
1DCF
 
 | |
6ICJ
 
 | | Crystal structure of PPARgamma with compound BR102375K | | Descriptor: | 2-butyl-5-[(3-tert-butyl-1,2,4-oxadiazol-5-yl)methyl]-6-methyl-3-{[2'-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)[1,1'-biphenyl]-4-yl]methyl}pyrimidin-4(3H)-one, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Hong, E, Chin, J, Jang, T.H, Kim, K.H, Jung, W, Kim, S.H. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structure of PPARgamma with compound BR102375K
To Be Published
|
|
2MJP
 
 | | STRUCTURE-BASED IDENTIFICATION OF THE BIOCHEMICAL FUNCTION OF A HYPOTHETICAL PROTEIN FROM METHANOCOCCUS JANNASCHII:MJ0226 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYROPHOSPHATASE | | Authors: | Hwang, K.Y, Chung, J.H, Han, Y.S, Kim, S.H, Cho, Y, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based identification of a novel NTPase from Methanococcus jannaschii.
Nat.Struct.Biol., 6, 1999
|
|
1XKA
 
 | |
1XKB
 
 | |
1KRL
 
 | | Crystal Structure of Racemic DL-monellin in P-1 | | Descriptor: | MONELLIN, CHAIN A, CHAIN B | | Authors: | Hung, L.W, Kohmura, M, Ariyoshi, Y, Kim, S.H. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural differences in D and L-monellin in the crystals of racemic mixture.
J.Mol.Biol., 285, 1999
|
|
5EY2
 
 | | Crystal structure of CodY from Bacillus cereus | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY | | Authors: | Han, A, Lee, W.C, Son, J, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
