6K8W
 
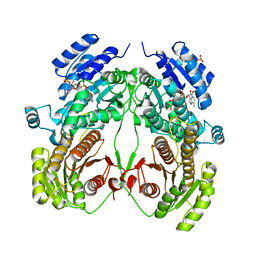 | | Crystal structure of N-domain with NADP of baterial malonyl-CoA reductase | | Descriptor: | NAD-dependent epimerase/dehydratase:Short-chain dehydrogenase/reductase SDR, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural insight into bi-functional malonyl-CoA reductase.
Environ.Microbiol., 22, 2020
|
|
6K8S
 
 | |
6KIE
 
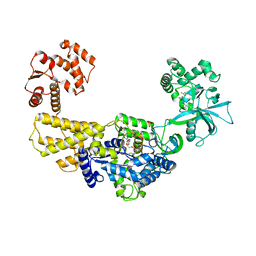 | | Crystal structure of human leucyl-tRNA synthetase, Leu-AMS-bound form | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-07-18 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
8WEQ
 
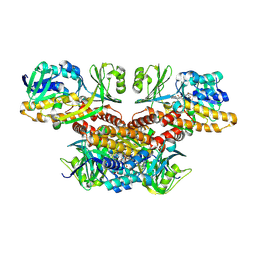 | | p-hydroxybenzoate 3-monooxygenase | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kim, S.B, Park, H.H. | | Deposit date: | 2023-09-18 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of p-hydroxybenzoate 3-monooxygenase
To Be Published
|
|
6K8V
 
 | |
6KR7
 
 | | Crystal structure of methylated human leucyl-tRNA synthetase, Leu-AMS-bound form | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE, Leucine--tRNA ligase, ... | | Authors: | Kim, S, Son, J, Kim, S, Hwang, K.Y. | | Deposit date: | 2019-08-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Leucine-sensing mechanism of leucyl-tRNA synthetase 1 for mTORC1 activation.
Cell Rep, 35, 2021
|
|
1YZ4
 
 | | Crystal structure of DUSP15 | | Descriptor: | SULFATE ION, dual specificity phosphatase-like 15 isoform a, octyl beta-D-glucopyranoside | | Authors: | Kim, S.J, Ryu, S.E, Jeong, D.G, Yoon, T.S, Kim, J.H, Cho, Y.H, Jeong, S.K, Lee, J.W, Son, J.H. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the catalytic domain of human VHY, a dual-specificity protein phosphatase
Proteins, 61, 2005
|
|
1AIN
 
 | |
2YYL
 
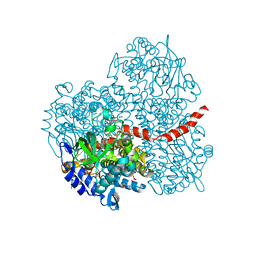 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) complexed with FAD | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
2GWO
 
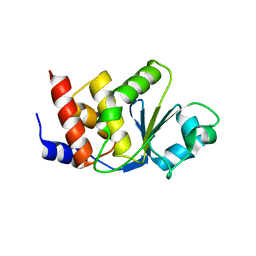 | | crystal structure of TMDP | | Descriptor: | Dual specificity protein phosphatase 13 | | Authors: | Kim, S.J, Ryu, S.E, Kim, J.H. | | Deposit date: | 2006-05-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human TMDP, a testis-specific dual specificity protein phosphatase: implications for substrate specificity
Proteins, 66, 2007
|
|
2LIG
 
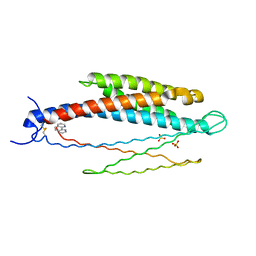 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR, ASPARTIC ACID, ... | | Authors: | Kim, S.-H, Yeh, J.I, Prive, G.G, Milburn, M, Scott, W, Koshland Junior, D.E. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
1THI
 
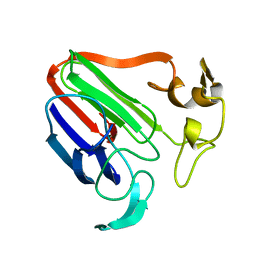 | |
7E43
 
 | |
6K8T
 
 | |
6K8U
 
 | |
2QDF
 
 | | Structure of N-terminal domain of E. Coli YaeT | | Descriptor: | MAGNESIUM ION, Outer membrane protein assembly factor yaeT | | Authors: | Kim, S, Malinverni, J.C, Sliz, P, Silhavy, T.J, Harrison, S.C, Kahne, D. | | Deposit date: | 2007-06-20 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of an essential component of the outer membrane protein assembly machine.
Science, 317, 2007
|
|
2G6Z
 
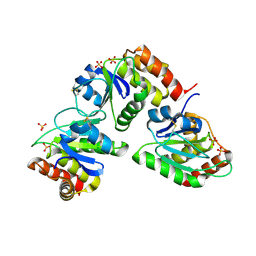 | | Crystal structure of human DUSP5 | | Descriptor: | Dual specificity protein phosphatase 5, SULFATE ION | | Authors: | Kim, S.J, Ryu, S.E. | | Deposit date: | 2006-02-26 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the catalytic domain of human DUSP5, a dual specificity MAP kinase protein phosphatase
Proteins, 66, 2007
|
|
4YED
 
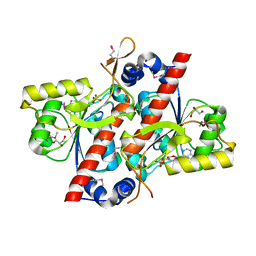 | | TcdA (CsdL) | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, tRNA threonylcarbamoyladenosine dehydratase | | Authors: | Kim, S, Park, S.Y. | | Deposit date: | 2015-02-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Escherichia coli TcdA (Also Known As CsdL) Reveals a Novel Topology and Provides Insight into the tRNA Binding Surface Required for N(6)-Threonylcarbamoyladenosine Dehydratase Activity
J.Mol.Biol., 427, 2015
|
|
8XJE
 
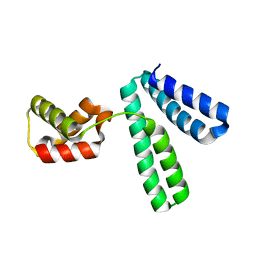 | |
8XJG
 
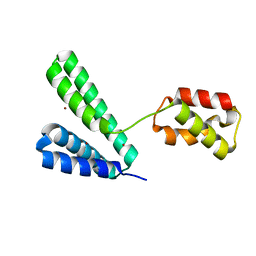 | |
2ED4
 
 | | Crystal structure of flavin reductase HpaC complexed with FAD and NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenae | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
1LAH
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
2ECU
 
 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2QCZ
 
 | | Structure of N-terminal domain of E. Coli YaeT | | Descriptor: | Outer membrane protein assembly factor yaeT | | Authors: | Kim, S, Malinverni, J.C, Sliz, P, Silhavy, T.J, Harrison, S.C, Kahne, D. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of an essential component of the outer membrane protein assembly machine.
Science, 317, 2007
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
