3KL9
 
 | | Crystal structure of PepA from Streptococcus pneumoniae | | Descriptor: | Glutamyl aminopeptidase, ZINC ION | | Authors: | Kim, K.K, Lee, S, Kim, D. | | Deposit date: | 2009-11-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the substrate specificity of PepA from Streptococcus pneumoniae, a dodecameric tetrahedral protease
Biochem.Biophys.Res.Commun., 391, 2010
|
|
1QU7
 
 | |
2EIF
 
 | | Eukaryotic translation initiation factor 5A from Methanococcus jannaschii | | Descriptor: | PROTEIN (EUKARYOTIC TRANSLATION INITIATION FACTOR 5A) | | Authors: | Kim, K.K, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1SHS
 
 | |
1EIF
 
 | | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A FROM METHANOCOCCUS JANNASCHII | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A | | Authors: | Kim, K.K, Hung, L.W, Yokota, H, Kim, R, Kim, S.H. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1OIL
 
 | | STRUCTURE OF LIPASE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 1996-12-06 | | Release date: | 1997-05-15 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
1KXU
 
 | | CYCLIN H, A POSITIVE REGULATORY SUBUNIT OF CDK ACTIVATING KINASE | | Descriptor: | CYCLIN H | | Authors: | Kim, K.K, Chamberin, H.M, Morgan, D.O, Kim, S.-H. | | Deposit date: | 1996-08-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase.
Nat.Struct.Biol., 3, 1996
|
|
1AUO
 
 | | CARBOXYLESTERASE FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | CARBOXYLESTERASE | | Authors: | Kim, K.K, Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of carboxylesterase from Pseudomonas fluorescens, an alpha/beta hydrolase with broad substrate specificity.
Structure, 5, 1997
|
|
4KIA
 
 | | Crystal structure of LmHde, heme-degrading enzyme, from Listeria monocytogenes | | Descriptor: | Lmo2213 protein | | Authors: | Kim, K.K, Duong, T, Kim, T. | | Deposit date: | 2013-05-02 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional characterization of an Isd-type haem-degradation enzyme from Listeria monocytogenes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1AUR
 
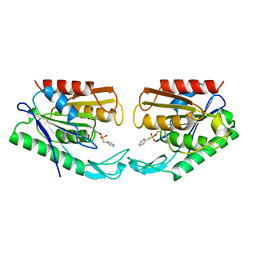 | |
5ZU1
 
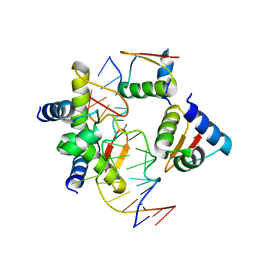 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*AP*GP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*TP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-05 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5ZUO
 
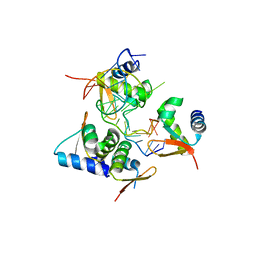 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5ZUP
 
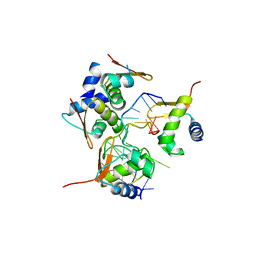 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*AP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5J6X
 
 | |
2ACJ
 
 | | Crystal structure of the B/Z junction containing DNA bound to Z-DNA binding proteins | | Descriptor: | 5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*GP*GP*CP*GP*CP*GP*CP*G)-3', 5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*AP*TP*AP*AP*AP*CP*C)-3', Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Lowenhaupt, K, Rich, A, Kim, Y.-G, Kim, K.K. | | Deposit date: | 2005-07-19 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases.
Nature, 437, 2005
|
|
5JNM
 
 | | Crystal structure of MtlD from Staphylococcus aureus at 1.7-Angstrom resolution | | Descriptor: | Mannitol-1-phosphate 5-dehydrogenase, SULFATE ION | | Authors: | Ta, H.M, Nguyen, T, Kim, T, Kim, K.K. | | Deposit date: | 2016-04-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Targeting Mannitol Metabolism as an Alternative Antimicrobial Strategy Based on the Structure-Function Study of Mannitol-1-Phosphate Dehydrogenase in Staphylococcus aureus.
Mbio, 10, 2019
|
|
5GUY
 
 | | Structural and functional study of chuY from E. coli CFT073 strain | | Descriptor: | Uncharacterized protein chuY | | Authors: | Kim, H, Choi, J, HA, S.C, Chaurasia, A.K, Kim, D, Kim, K.K. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural and functional study of ChuY from Escherichia coli strain CFT073
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5HOE
 
 | | Crystal structrue of Est24, a carbohydrate acetylesterase from Sinorhizobium meliloti | | Descriptor: | HEXAETHYLENE GLYCOL, Hydrolase, PHOSPHATE ION | | Authors: | Oh, C, Ryu, B.H, An, D.R, Nguyen, D.D, Yoo, W, Kim, T, Ngo, D.T, Kim, H.S, Park, J.S, Kim, K.K, Kim, T.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of an Octameric Carbohydrate Acetylesterase from Sinorhizobium meliloti.
Febs Lett., 590, 2016
|
|
3BG4
 
 | | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Kim, H, Chu, T.T.T, Kim, D.Y, Kim, D.R, Nguyen, C.M.T, Choi, J, Lee, J.R, Hahn, M.J, Kim, K.K. | | Deposit date: | 2007-11-26 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor.
J.Mol.Biol., 376, 2008
|
|
5C5Z
 
 | | Crystal structure analysis of c4763, a uropathogenic E. coli-specific protein | | Descriptor: | Glutamyl-tRNA amidotransferase | | Authors: | Kim, H, Choi, J, Kim, D, Kim, K.K. | | Deposit date: | 2015-06-22 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure analysis of c4763, a uropathogenic Escherichia coli-specific protein.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1BCC
 
 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
8WP9
 
 | | Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5 | | Descriptor: | Small heat shock protein HSP16.5 | | Authors: | Lee, J, Ryu, B, Kim, T, Kim, K.K. | | Deposit date: | 2023-10-09 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of a 16.5-kDa small heat-shock protein from Methanocaldococcus jannaschii.
Int.J.Biol.Macromol., 258, 2024
|
|
2BCC
 
 | | STIGMATELLIN-BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 1998-09-18 | | Release date: | 1999-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
1YGZ
 
 | | Crystal Structure of Inorganic Pyrophosphatase from Helicobacter pylori | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Wu, C.A, Lokanath, N.K, Kim, D.Y, Park, H.J, Hwang, H.Y, Kim, S.T, Suh, S.W, Kim, K.K. | | Deposit date: | 2005-01-06 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of inorganic pyrophosphatase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5GKV
 
 | | Crystal Structure of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus | | Descriptor: | Esterase A | | Authors: | Ngo, T.D, Ryu, B.H, Kim, B.Y, Yoo, W.K, Lee, E.J, Lee, S.J, Kim, T.D, Kim, K.K. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Biochemical and Structural Analysis of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus
To Be Published
|
|
