1RUH
 
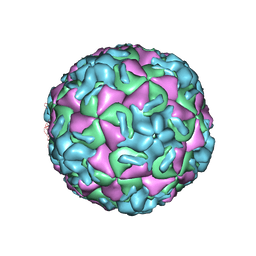 | | RHINOVIRUS 14 MUTANT N1219S COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52084 | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1Q1G
 
 | | Crystal structure of Plasmodium falciparum PNP with 5'-methylthio-immucillin-H | | Descriptor: | 3,4-DIHYDROXY-2-[(METHYLSULFANYL)METHYL]-5-(4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)PYRROLIDINIUM, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Shi, W, Ting, L.M, Kicska, G.A, Lewandowicz, A, Tyler, P.C, Evans, G.B, Furneaux, R.H, Kim, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-07-19 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Plasmodium falciparum Purine Nucleoside Phosphorylase: CRYSTAL STRUCTURES, IMMUCILLIN INHIBITORS, AND DUAL CATALYTIC FUNCTION.
J.Biol.Chem., 279, 2004
|
|
6IHE
 
 | |
7CBE
 
 | |
6IUM
 
 | |
6IUN
 
 | |
6IJK
 
 | |
6IJ4
 
 | |
6J2U
 
 | |
6IHD
 
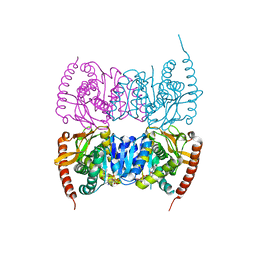 | | Crystal structure of Malate dehydrogenase from Metallosphaera sedula | | Descriptor: | 2-ETHOXYETHANOL, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, D, Kim, K.J. | | Deposit date: | 2018-09-29 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and biochemical characterization of malate dehydrogenase from Metallosphaera sedula
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IJ3
 
 | |
6IJ5
 
 | |
6KUS
 
 | |
6KUQ
 
 | |
6L3O
 
 | |
6IJ6
 
 | |
6KUO
 
 | |
4NE2
 
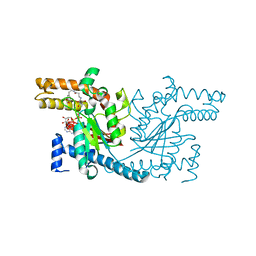 | | Pantothenamide-bound Pantothenate Kinase from Klebsiella pneumoniae | | Descriptor: | (R)-N-(3-((2-(benzo[d][1,3]dioxol-5-yl)ethyl)amino)-3-oxopropyl)-2,4-dihydroxy-3,3-dimethylbutanamide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Kim, K.P, Smil, D, Park, H.W. | | Deposit date: | 2013-10-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a new N-substituted pantothenamide bound to pantothenate kinases from Klebsiella pneumoniae and Staphylococcus aureus.
Proteins, 82, 2014
|
|
7FBG
 
 | |
5Z7R
 
 | |
5ZWR
 
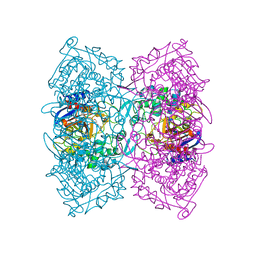 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, Est-Y29, GLYCEROL | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWQ
 
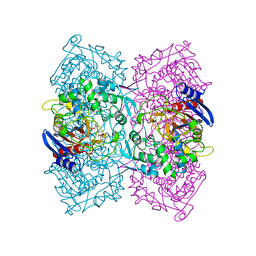 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29, GLYCEROL, ethyl (2S)-2-[3-(benzenecarbonyl)phenyl]propanoate | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZRD
 
 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at low pH condition | | Descriptor: | CITRIC ACID, COPPER (II) ION, GLYCEROL, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
5ZRE
 
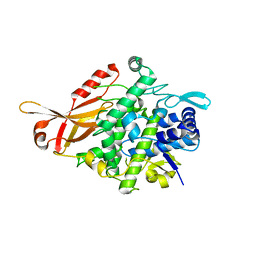 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at high pH condition | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN ATOM, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
5YS9
 
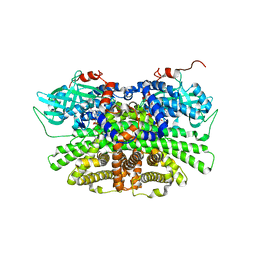 | | Crystal structure of acyl-coA oxidase3 from Yarrowia lipolytica | | Descriptor: | Acyl-coenzyme A oxidase 3, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2017-11-13 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Acyl-CoA Oxidase 3 fromYarrowia lipolyticawith Specificity for Short-Chain Acyl-CoA.
J. Microbiol. Biotechnol., 28, 2018
|
|
