4KU7
 
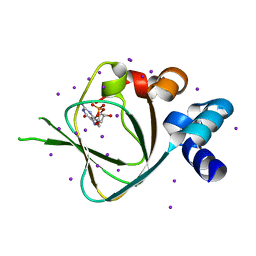 | | Structures of PKGI Reveal a cGMP-Selective Activation Mechanism | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, IODIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Huang, G.Y, Kim, J.J, Reger, A.S, Lorenz, R, Moon, E.W, Casteel, D.E, Sankaran, B, Herberg, F.W, Kim, C. | | Deposit date: | 2013-05-21 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Cyclic-Nucleotide Selectivity and cGMP-Selective Activation of PKG I.
Structure, 22, 2014
|
|
7BVB
 
 | |
7BVA
 
 | |
5I76
 
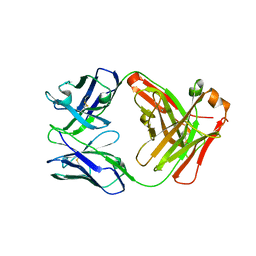 | | Crystal structure of FM318, a recombinant Fab adopted from cetuximab | | Descriptor: | FM318_heavy_cahin, FM318_light_chain | | Authors: | Sim, D.W, Kim, J.H, Seok, S.H, Seo, M.D, Kim, Y.P, Won, H.S. | | Deposit date: | 2016-02-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Bacterial production and structure-functional validation of a recombinant antigen-binding fragment (Fab) of an anti-cancer therapeutic antibody targeting epidermal growth factor receptor.
Appl.Microbiol.Biotechnol., 100, 2016
|
|
6KQB
 
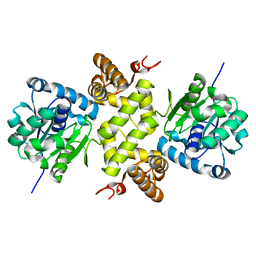 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
6KQ9
 
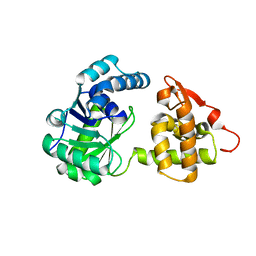 | | A long chain secondary alcohol dehydrogenase of Micrococcus luteus | | Descriptor: | 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Kim, H.J, Kim, J.S. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Cofactor specificity engineering of a long-chain secondary alcohol dehydrogenase from Micrococcus luteus for redox-neutral biotransformation of fatty acids.
Chem.Commun.(Camb.), 55, 2019
|
|
3UFC
 
 | |
4O9C
 
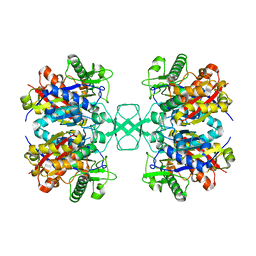 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
6XAI
 
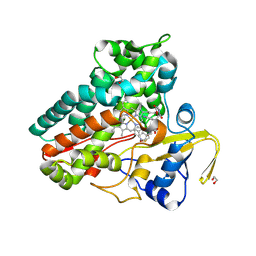 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-Pro) | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, NzeB, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6XAL
 
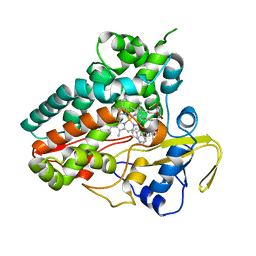 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-Val) | | Descriptor: | (3S,6S)-3-[(1H-indol-3-yl)methyl]-6-(propan-2-yl)piperazine-2,5-dione, NzeB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6XAK
 
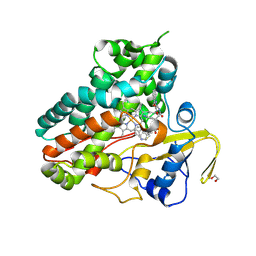 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-Pro) and cyclo-(L-Trp-L-Trp) | | Descriptor: | (3S,6S)-3,6-bis[(1H-indol-3-yl)methyl]piperazine-2,5-dione, (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6XAM
 
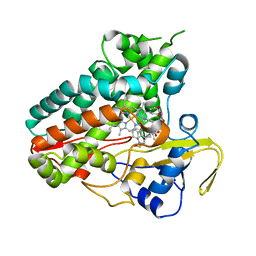 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-homoalanine) | | Descriptor: | (3S,6S)-3-ethyl-6-[(1H-indol-3-yl)methyl]piperazine-2,5-dione, 1,2-ETHANEDIOL, NzeB, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6XAJ
 
 | | Crystal structure of NzeB | | Descriptor: | 1,2-ETHANEDIOL, NzeB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6K65
 
 | | Application of anti-helix antibodies in protein structure determination (9014-1P4B) | | Descriptor: | 1P4B variable heavy chain, 1P4B variable light chain, Immunoglobulin G-binding protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6B
 
 | | Application of anti-helix antibodies in protein structure determination (8496-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K3M
 
 | | Application of anti-helix antibodies in protein structure determination (8189-3LRH) | | Descriptor: | 3LRH intrabody, SpA IgG-binding domain protein,Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7V5C
 
 | | Cryo-EM structure of the mouse ABCB9 (ADP.BeF3-bound) | | Descriptor: | ABC-type oligopeptide transporter ABCB9, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Kim, J.W, Ju, S, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
7V5D
 
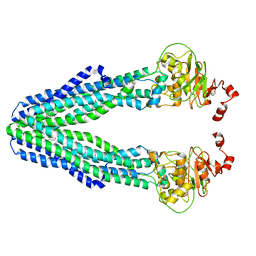 | | Cryo-EM structure of the mouse ABCB9 (PG-bound) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ABC-type oligopeptide transporter ABCB9 | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Ju, S, Kim, J.W, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
6K67
 
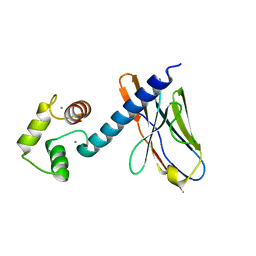 | | Application of anti-helix antibodies in protein structure determination (9011-3LRH) | | Descriptor: | 3LRH introbody, CALCIUM ION, Engineered calmodulin | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5H62
 
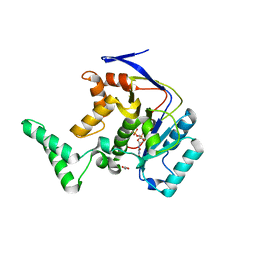 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Transferase, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
6KMA
 
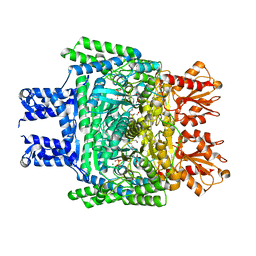 | | Crystal structure of SucA with glycolaldehyde-1-13C from Vibrio vulnificus | | Descriptor: | 2-oxidanylethanal, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
4Y0M
 
 | |
5H63
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | MANGANESE (II) ION, Transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
4Q6Q
 
 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6K68
 
 | | Application of anti-helix antibodies in protein structure determination (8420-3MNZ) | | Descriptor: | 3MNZ Variable heavy chain, 3MNZ Variable light chain, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
