2DS8
 
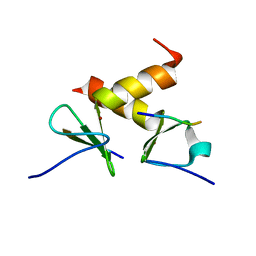 | | Structure of the ZBD-XB complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
3A3W
 
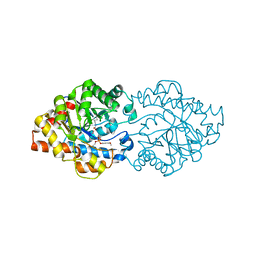 | | Structure of OpdA mutant (G60A/A80V/S92A/R118Q/K185R/Q206P/D208G/I260T/G273S) with diethyl 4-methoxyphenyl phosphate bound in the active site | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
3A3X
 
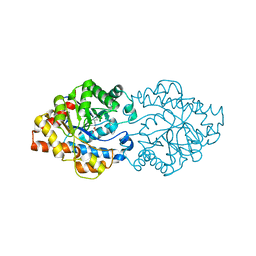 | | Structure of OpdA mutant (G60A/A80V/R118Q/K185R/Q206P/D208G/I260T/G273S) | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
4R27
 
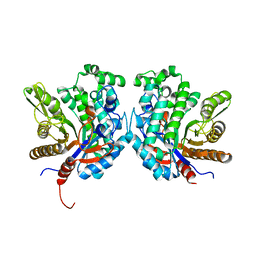 | | Crystal structure of beta-glycosidase BGL167 | | Descriptor: | Glycoside hydrolase | | Authors: | Park, S.J, Choi, J.M, Kyeong, H.H, Kim, S.G, Kim, H.S. | | Deposit date: | 2014-08-09 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational design of a beta-glycosidase with high regiospecificity for triterpenoid tailoring
Chembiochem, 16, 2015
|
|
4HAB
 
 | | Crystal structure of Plk1 Polo-box domain in complex with PL-49 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION, PL-49, ... | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-04-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of cyclic peptomer inhibitors targeting the polo-box domain of polo-like kinase 1.
Bioorg.Med.Chem., 21, 2013
|
|
5XWB
 
 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
1QBY
 
 | | THE SOLUTION STRUCTURE OF A BAY-REGION 1R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*GP*GP*AP*CP*(BZA)AP*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP* G)-3' | | Authors: | Li, Z, Mao, H, Kim, H.-Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-04-27 | | Release date: | 1999-05-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (-)-(1R,2S,3R, 4S)-N6-[1-benz[a]anthracenyl]-2'-deoxyadenosyl adduct in an oligodeoxynucleotide containing the human N-ras codon 61 sequence.
Biochemistry, 38, 1999
|
|
6LSY
 
 | | AAA+ ATPase, ClpL from Streptococcus pneumoniae - ATP bound | | Descriptor: | ATP-dependent Clp protease, ATP-binding subunit | | Authors: | Kim, G, Lee, S.G, Han, S, Jung, J, Jeong, H.S, Hyun, J.K, Rhee, D.K, Kim, H.M, Lee, S. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.33 Å) | | Cite: | ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Faseb J., 34, 2020
|
|
3X2Y
 
 | | Crystal structure of metallo-beta-lactamase H8A from Thermotoga maritima | | Descriptor: | NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of metallo-beta-lactamase H8A from Thermotoga maritima
To be Published
|
|
3X30
 
 | | Crystal structure of metallo-beta-lactamase from Thermotoga maritima | | Descriptor: | MANGANESE (II) ION, NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Crystal structure of metallo-beta-lactamase from Thermotoga maritima
To be Published
|
|
3X2X
 
 | | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima | | Descriptor: | MANGANESE (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima
To be Published
|
|
3X2Z
 
 | | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima | | Descriptor: | NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima
To be Published
|
|
3UYV
 
 | | Crystal structure of a glycosylated ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
3UYU
 
 | | Structural basis for the antifreeze activity of an ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, GLYCEROL | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
7KPW
 
 | | Structure of the H-lobe of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12 | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
1YUM
 
 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | 'Probable nicotinate-nucleotide adenylyltransferase, CITRIC ACID, NICOTINATE MONONUCLEOTIDE | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
1UAK
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAL
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
3GN4
 
 | | Myosin lever arm | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Mukherjea, M, Llinas, P, Kim, H, Travaglia, M, Safer, D, Zong, A.B, Menetrey, J, Franzini-Armstrong, C, Selvin, P.R, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach
Mol.Cell, 35, 2009
|
|
2A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
4HY2
 
 | |
3DCM
 
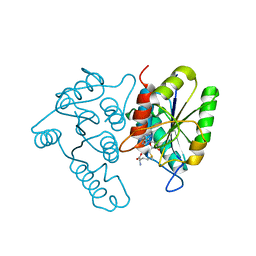 | | Crystal structure of the Thermotoga maritima SPOUT family RNA-methyltransferase protein Tm1570 in complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein TM_1570 | | Authors: | Kim, D.J, Kim, H.S, Lee, S.J, Suh, S.W. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Thermotoga maritima SPOUT superfamily RNA methyltransferase Tm1570 in complex with S-adenosyl-L-methionine
Proteins, 74, 2009
|
|
8I9J
 
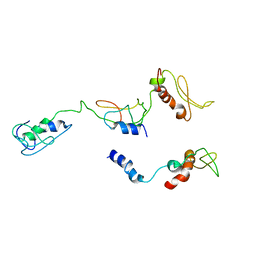 | | The PKR and E3L complex | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase, RNA-binding protein E3 | | Authors: | Han, C.W, Kim, H.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (6.39 Å) | | Cite: | Structural study of novel vaccinia virus E3L and dsRNA-dependent protein kinase complex.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
