7D4V
 
 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase Y91F/C120S variant | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7CSM
 
 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase C120S variant | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-08-15 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7CDC
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRP peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDF
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRK peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDG
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDD
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDE
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRKR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
7EIF
 
 | | Crystal structure of GAS41 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 4 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | GAS41 promotes H2A.Z deposition through recognition of the N terminus of histone H3 by the YEATS domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7FHS
 
 | | Crystal structure of DYRK1A in complex with RD0392 | | Descriptor: | (5~{Z})-5-[(3-ethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL | | Authors: | Kikuchi, M, Sumida, T, Hosoya, T, Kii, I, Umehara, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Eur.J.Med.Chem., 227, 2022
|
|
7FHT
 
 | | Crystal structure of DYRK1A in complex with RD0448 | | Descriptor: | (5~{Z})-5-[(3-ethynyl-4-methoxy-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Kikuchi, M, Sumida, Y, Hosoya, T, Kii, I, Umehara, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Eur.J.Med.Chem., 227, 2022
|
|
1QSW
 
 | |
6AIG
 
 | |
1LHJ
 
 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
1LHI
 
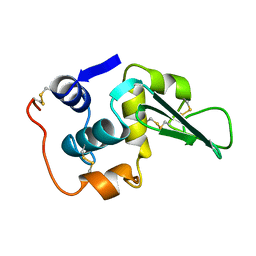 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
1LHK
 
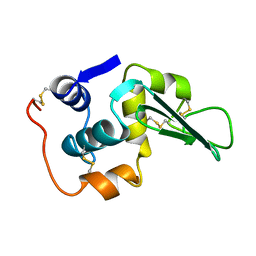 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
1LHH
 
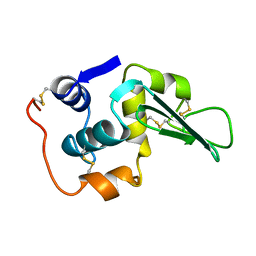 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
1LHL
 
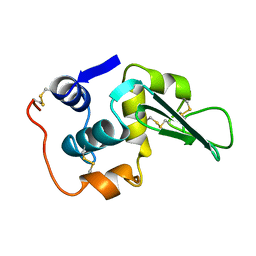 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
1YAM
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAN
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAQ
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAO
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAP
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1LZ5
 
 | | STRUCTURAL AND FUNCTIONAL ANALYSES OF THE ARG-GLY-ASP SEQUENCE INTRODUCED INTO HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, HUMAN LYSOZYME | | Authors: | Matsushima, M, Inaka, K, Yamada, T, Sekiguchi, K, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme.
J.Biol.Chem., 268, 1993
|
|
1LZ4
 
 | | ENTHALPIC DESTABILIZATION OF A MUTANT HUMAN LYSOZYME LACKING A DISULFIDE BRIDGE BETWEEN CYSTEINE-77 AND CYSTEINE-95 | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Kuroki, R, Taniyama, Y, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enthalpic destabilization of a mutant human lysozyme lacking a disulfide bridge between cysteine-77 and cysteine-95.
Biochemistry, 31, 1992
|
|
