1R2N
 
 | | NMR structure of the all-trans retinal in dark-adapted Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, terLaak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-09-29 | | Release date: | 2003-10-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4FJV
 
 | | Crystal Structure of Human Otubain2 and Ubiquitin Complex | | Descriptor: | ETHANAMINE, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Altun, M, Walter, T.S, Kramer, H.B, Iphofer, A, David, Y, Komsany, A, Ternette, N, Nicholson, B, Navon, A, Stuart, D.I, Ren, J, Kessler, B.M. | | Deposit date: | 2012-06-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The human otubain2-ubiquitin structure provides insights into the cleavage specificity of poly-ubiquitin-linkages.
Plos One, 10, 2015
|
|
2VKZ
 
 | | Structure of the cerulenin-inhibited fungal fatty acid synthase type I multienzyme complex | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FATTY ACID SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Johansson, P, Wiltschi, B, Kumari, P, Kessler, B, Vonrhein, C, Vonck, J, Oesterhelt, D, Grininger, M. | | Deposit date: | 2008-01-07 | | Release date: | 2008-08-12 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Inhibition of the Fungal Fatty Acid Synthase Type I Multienzyme Complex.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7YZP
 
 | | Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA hairpin (59-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7YZO
 
 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7Z03
 
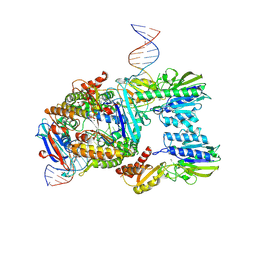 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (39-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
6S85
 
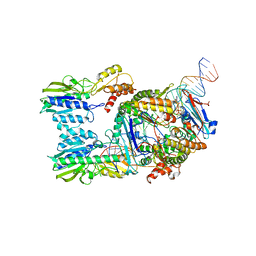 | | Cutting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), DNA (32-MER), ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
1R84
 
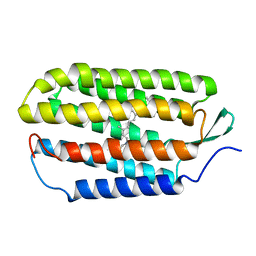 | | NMR structure of the 13-cis-15-syn retinal in dark_adapted bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, Ter Laak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6S6V
 
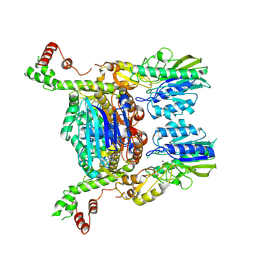 | | Resting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ATPgS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Nuclease SbcCD subunit C, ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-04 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
5DAC
 
 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with DNA | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
5NGF
 
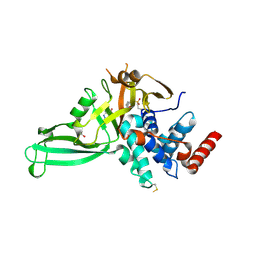 | | Crystal structure of USP7 in complex with the covalent inhibitor, FT827 | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[2-[4-[4-[(1-methyl-4-oxidanylidene-pyrazolo[3,4-d]pyrimidin-5-yl)methyl]-4-oxidanyl-piperidin-1-yl]carbonylphenyl]phenyl]ethanesulfonamide | | Authors: | Krajewski, W.W, Turnbull, A.P, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
5NGE
 
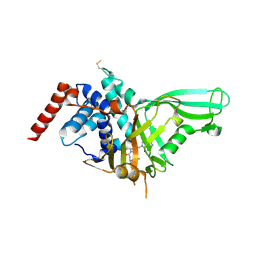 | | Crystal structure of USP7 in complex with the non-covalent inhibitor, FT671 | | Descriptor: | 5-[[1-[(3~{S})-4,4-bis(fluoranyl)-3-(3-fluoranylpyrazol-1-yl)butanoyl]-4-oxidanyl-piperidin-4-yl]methyl]-1-(4-fluorophenyl)pyrazolo[3,4-d]pyrimidin-4-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Turnbull, A.P, Krajewski, W.W, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
5DA9
 
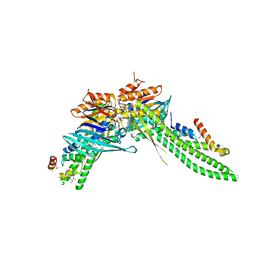 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with the Rad50-binding domain of Mre11 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative double-strand break protein, ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
1OFI
 
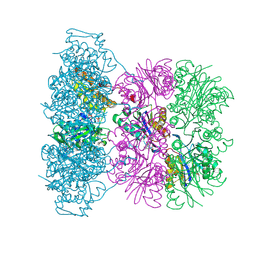 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1OFH
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
5QHV
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000299a | | Descriptor: | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QHZ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000385a | | Descriptor: | 2-cyano-~{N}-cyclohexyl-ethanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHY
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000462a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000506a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHU
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMSOA000341b | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-(2-hydroxyphenyl)acetamide, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI4
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000466a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QI0
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000352a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHW
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000347a | | Descriptor: | 2-methyl-~{N}-(2-methylpropyl)imidazo[1,2-a]pyridine-3-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI3
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000475a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI6
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000597a | | Descriptor: | 4-[(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)amino]phenol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
