7FBA
 
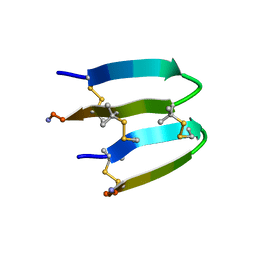 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd2 | | Descriptor: | ALA-LE1-CYS-GLU-CYS-GLY-PRO-THR-ARG-GLU-CYS-LYS-NH2, GLU-CYS-ARG-GLU-TYR-GLY-PRO-LE1-LYS-LE1-LE1-ALA-NH2 | | Authors: | Yao, H, Yao, S, Moyer, A, Zheng, Y, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
4JKY
 
 | |
4JLB
 
 | |
4JZK
 
 | |
4JLP
 
 | |
2V5R
 
 | | Structural basis for Dscam isoform specificity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DSCAM, GLYCEROL | | Authors: | Meijers, R, Puettmann-Holgado, R, Skiniotis, G, Liu, J.-H, Walz, T, Schmucker, D, Wang, J.-H. | | Deposit date: | 2007-07-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Dscam Isoform Specificity
Nature, 449, 2007
|
|
4JL5
 
 | |
2CMO
 
 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | Descriptor: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
7CBC
 
 | |
4JLO
 
 | |
2IVZ
 
 | | Structure of TolB in complex with a peptide of the colicin E9 T- domain | | Descriptor: | CALCIUM ION, COLICIN-E9, PROTEIN TOLB | | Authors: | Loftus, S.R, Walker, D, Mate, M.J, Bonsor, D.A, James, R, Moore, G.R, Kleanthous, C. | | Deposit date: | 2006-06-23 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Competitive Recruitment of the Periplasmic Translocation Portal Tolb by a Natively Disordered Domain of Colicin E9
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1OUR
 
 | | LecB (PA-LII) in complex with mannose | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
2JYZ
 
 | | CG7054 solution structure | | Descriptor: | CG7054-PA | | Authors: | Rautureau, G, Jouvensal, L, Vovelle, F, Schoentgen, F, Locker, D, Decoville, M, Damblon, C. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a phosphatidyl-ethanolamine binding protein from Drosophila
Proteins, 78, 2009
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
2JLS
 
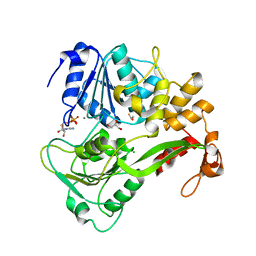 | | Dengue virus 4 NS3 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLU
 
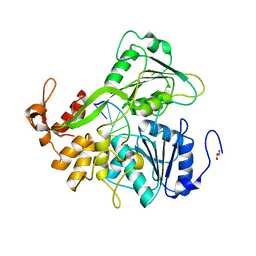 | | Dengue virus 4 NS3 helicase in complex with ssRNA | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', GLYCEROL, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2MSK
 
 | |
2JLX
 
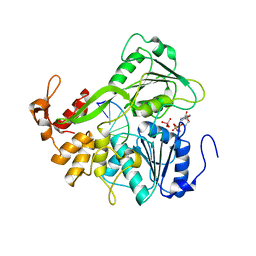 | | Dengue virus 4 NS3 helicase in complex with ssRNA and ADP-Vanadate | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLW
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA2 | | Descriptor: | 5'-R(*UP*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', GLYCEROL, PHOSPHATE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein
Embo J., 27, 2008
|
|
1OUX
 
 | | LecB (PA-LII) sugar-free | | Descriptor: | CALCIUM ION, SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OXC
 
 | | LecB (PA-LII) in complex with FUCOSE | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-04-02 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
4IAQ
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
1OVP
 
 | | LecB (PA-LII) in complex with fructose | | Descriptor: | CALCIUM ION, beta-D-fructopyranose, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-27 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
7FU4
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(2-fluoro-4-phenylphenyl)methylamino]-5-propyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-2-{[(3-fluoro[1,1'-biphenyl]-4-yl)methyl]amino}-5-propyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
1PUX
 
 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
