5G40
 
 | | Crystal structure of adenylate kinase ancestor 4 with Zn and AMP-ADP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G3Z
 
 | | Crystal structure of adenylate kinase ancestor 3 with Zn, Mg and Ap5A bound | | Descriptor: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
3Q9U
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | COENZYME A, CoA binding protein, consensus ankyrin repeat | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
2C8I
 
 | | Complex Of Echovirus Type 12 With Domains 1, 2, 3 and 4 Of Its Receptor Decay Accelerating Factor (Cd55) By Cryo Electron Microscopy At 16 A | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Pettigrew, D.M, Williams, D.T, Kerrigan, D, Evans, D.J, Lea, S.M, Bhella, D. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural and Functional Insights Into the Interaction of Echoviruses and Decay-Accelerating Factor.
J.Biol.Chem., 281, 2006
|
|
4ZHO
 
 | | The crystal structure of Arabidopsis ferredoxin 2 with 2Fe-2S cluster | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZHP
 
 | | The crystal structure of Potato ferredoxin I with 2Fe-2S cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Potato Ferredoxin I | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZGV
 
 | | The Crystal Structure of the Ferredoxin Receptor FusA from Pectobacterium atrosepticum SCRI1043 | | Descriptor: | Ferredoxin receptor, LAURYL DIMETHYLAMINE-N-OXIDE, octyl beta-D-glucopyranoside | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-24 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
2MRA
 
 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459 | | Descriptor: | De novo designed protein OR459 | | Authors: | Pulavarti, S.V.S.R.K, Kipnis, Y, Sukumaran, D, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459
To be Published
|
|
5IYT
 
 | | Complex structure of EV-B93 main protease 3C with N-Ethyl 4-((1-cycloheptyl-1,2-dihydropyrazol-3-one-5-yl)-amino)-4-oxo-2Z-butenamide | | Descriptor: | EV-B93 main protease 3C, N-Ethyl 4-((1-cycloheptyl-1,2-dihydropyrazol-3-one-5-yl)-amino)-4-oxo-butanamide | | Authors: | Kaczmarska, Z, Becker, D, Rademann, J, Coll, M. | | Deposit date: | 2016-03-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding fragments.
Nat Commun, 7, 2016
|
|
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
5G3Y
 
 | | Crystal structure of adenylate kinase ancestor 1 with Zn and ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ZINC ION | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5A6N
 
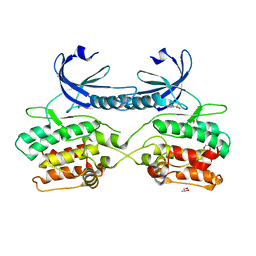 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 | | Descriptor: | 5-(3-SULFAMOYLPHENYL)-1H-1,2,3,4-TETRAZOL-1-IDE, DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, ... | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5G41
 
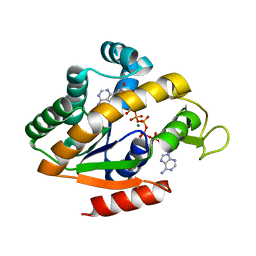 | | Crystal structure of adenylate kinase ancestor 4 with Zn, Mg and Ap5A bound | | Descriptor: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
7G4R
 
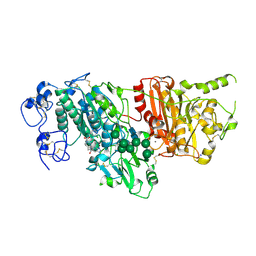 | | Crystal Structure of rat Autotaxin in complex with (3-chloro-5-methylsulfonylphenyl)methyl rac-(3aR,8aS)-2-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,5,7,8,8a-octahydropyrrolo[3,4-d]azepine-6-carboxylate, i.e. SMILES N1(CC[C@@H]2[C@H](CC1)CN(C2)C(=O)c1ccc2c(c1)N=NN2)C(=O)OCc1cc(cc(c1)Cl)S(=O)(=O)C with IC50=0.00849941 microM | | Descriptor: | CALCIUM ION, CHLORIDE ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G44
 
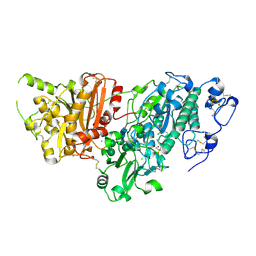 | | Crystal Structure of rat Autotaxin in complex with (3,5-dichlorophenyl)methyl rac-(3aR,6aS)-2-(4-sulfamoylbenzoyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrole-5-carboxylate, i.e. SMILES C1N(C[C@H]2[C@@H]1CN(C2)C(=O)c1ccc(cc1)S(=O)(=O)N)C(=O)OCc1cc(cc(c1)Cl)Cl with IC50=0.0573624 microM | | Descriptor: | (3,5-dichlorophenyl)methyl (3aS,6aS)-5-(4-sulfamoylbenzoyl)hexahydropyrrolo[3,4-c]pyrrole-2(1H)-carboxylate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G49
 
 | | Crystal Structure of rat Autotaxin in complex with 8-(2-fluorophenyl)-4-[3-(5-methyl-1,3,4-oxadiazol-2-yl)phenyl]-1,3-dihydro-1,5-benzodiazepin-2-one | | Descriptor: | (4M,8P)-8-(2-fluorophenyl)-4-[(3P)-3-(5-methyl-1,3,4-oxadiazol-2-yl)phenyl]-1,3-dihydro-2H-1,5-benzodiazepin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Goetschi, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G5F
 
 | | Crystal Structure of rat Autotaxin in complex with 1H-benzotriazol-5-yl-[rac-(1R,2R,6S,7S)-9-[4-(cyclopropylmethoxy)naphthalene-2-carbonyl]-4,9-diazatricyclo[5.3.0.02,6]decan-4-yl]methanone, i.e. SMILES N1(C[C@H]2[C@@H](C1)[C@H]1[C@@H]2CN(C1)C(=O)c1ccc2c(c1)N=NN2)C(=O)c1cc(c2c(c1)cccc2)OCC1CC1 with IC50=0.00985223 microM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G4A
 
 | | Crystal Structure of rat Autotaxin in complex with (3-chloro-5-methylsulfonylphenyl)methyl 2-(1H-benzotriazole-5-carbonyl)-2,7-diazaspiro[3.5]nonane-7-carboxylate, i.e. SMILES c1c(cc(cc1S(=O)(=O)C)COC(=O)N1CCC2(CC1)CN(C2)C(=O)c1ccc2c(c1)N=NN2)Cl with IC50=0.332347 microM | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G51
 
 | | Crystal Structure of rat Autotaxin in complex with (5Z)-5-[(4-phenoxyphenyl)methylidene]-2-pyrrolidin-1-yl-1,3-thiazol-4-one, i.e. SMILES C1(=Cc2ccc(Oc3ccccc3)cc2)/C(=O)N=C(S1)N1CCCC1 with IC50=0.135695 microM | | Descriptor: | (5Z)-5-[(4-phenoxyphenyl)methylidene]-2-(pyrrolidin-1-yl)-1,3-thiazol-4(5H)-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Chen, S, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G55
 
 | | Crystal Structure of rat Autotaxin in complex with [3-chloro-5-(trifluoromethoxy)phenyl]methyl rac-(3aR,6aS)-2-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrole-5-carboxylate, i.e. SMILES C1N(C[C@@H]2[C@H]1CN(C2)C(=O)c1ccc2c(c1)N=NN2)C(=O)OCc1cc(cc(c1)Cl)OC(F)(F)F with IC50=0.00942633 microM | | Descriptor: | ACETATE ION, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G4E
 
 | | Crystal Structure of rat Autotaxin in complex with (5E)-5-[(4-tert-butylphenyl)methylidene]-2-pyrrolidin-1-yl-1,3-thiazol-4-one, i.e. SMILES N1=C(S/C(=C/c2ccc(C(C)(C)C)cc2)/C1=O)N1CCCC1 with IC50=0.210793 microM | | Descriptor: | (5Z)-5-[(4-tert-butylphenyl)methylidene]-2-(pyrrolidin-1-yl)-1,3-thiazol-4(5H)-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G5G
 
 | | Crystal Structure of rat Autotaxin in complex with 1-[2-(cyclohexylmethyl)-1,3-dioxoisoindol-4-yl]-N-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]piperidine-4-carboxamide, i.e. SMILES N1(C(=O)c2c(C1=O)cccc2N1CCC(C(=O)NCC2=NON=C2C)CC1)CC1CCCCC1 with IC50=0.0435744 microM | | Descriptor: | 1-[2-(cyclohexylmethyl)-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl]-N-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]piperidine-4-carboxamide, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G73
 
 | | Crystal Structure of rat Autotaxin in complex with 6-(4-acetylpiperazin-1-yl)-3-[[1-[(3,4-dichlorophenyl)methyl]triazol-4-yl]methyl]quinazolin-4-one, i.e. SMILES c1(ccc2c(c1)C(=O)N(C=N2)CC1=CN(N=N1)Cc1ccc(c(c1)Cl)Cl)N1CCN(CC1)C(=O)C with IC50=0.0309414 microM | | Descriptor: | 6-(4-acetylpiperazin-1-yl)-3-({1-[(3,4-dichlorophenyl)methyl]-1H-1,2,3-triazol-4-yl}methyl)quinazolin-4(3H)-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Martin-Rainer, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
5A6O
 
 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4K46
 
 | | Crystal Structure of Adenylate Kinase from Photobacterium profundum | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, ... | | Authors: | Cho, Y.-J, Kerns, S.J, Kern, D. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Alike but Different: Adenylate kinases from E. Coli, Aquifex, and P. profundum
To be Published
|
|
