7JHZ
 
 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[8] rotavirus strain BM13851 in complex with LNDFH I | | Descriptor: | GLYCEROL, Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Xu, S, Stuckert, M.R, McGinnis, K.R, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KHU
 
 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[4] rotavirus strain BM5265 in complex with LNDFH I | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Xu, S, Stuckert, M, Burnside, R, McGinnis, K, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KI5
 
 | | Crystal structure of P[6] rotavirus vp8* in complex with LNT | | Descriptor: | Capsid protein, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Xu, S, Kennedy, M.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2VH3
 
 | | ranasmurfin | | Descriptor: | GLYCEROL, RANASMURFIN, SULFATE ION, ... | | Authors: | Oke, M, Ching, R.T, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, Bloch Junior, C, Botting, C.H, Walsh, M.A, Latiff, A.A, Kennedy, M.W, Cooper, A, Naismith, J.H. | | Deposit date: | 2007-11-17 | | Release date: | 2007-12-04 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Unusual Chromophore and Cross-Links in Ranasmurfin: A Blue Protein from the Foam Nests of a Tropical Frog.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
2RN7
 
 | | NMR solution structure of TnpE protein from Shigella flexneri. Northeast Structural Genomics Target SfR125 | | Descriptor: | IS629 orfA | | Authors: | Ramelot, T.A, Cort, J.R, Semesi, A, Garcia, M, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-08 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of TnpE protein from Shigella flexneri. Northeast
Structural Genomics Target SfR125
To be Published
|
|
2WGO
 
 | | Structure of ranaspumin-2, a surfactant protein from the foam nests of a tropical frog | | Descriptor: | RANASPUMIN-2 | | Authors: | Mackenzie, C.D, Smith, B.O, Kennedy, M.W, Cooper, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Ranaspumin-2: Structure and Function of a Surfactant Protein from the Foam Nests of a Tropical Frog.
Biophys.J., 96, 2009
|
|
2AYJ
 
 | | Solution structure of 50S ribosomal protein L40e from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L40e, ZINC ION | | Authors: | Wu, B, Yee, A, Lukin, J, Lemak, A, Semesi, A, Ramelot, T, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-07 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus
Protein Sci., 17, 2008
|
|
3N90
 
 | | The 1.7 Angstrom resolution crystal structure of AT2G44920, a pentapeptide repeat protein from Arabidopsis thaliana thylakoid lumen. | | Descriptor: | SULFATE ION, Thylakoid lumenal 15 kDa protein 1, chloroplastic | | Authors: | Ni, S, Mckgookey, M, Tinch, S.L, Jones, A.N, Jayaraman, S, Kennedy, M.A. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 Angstrom resolution crystal structure of AT2G44920, a pentapeptide repeat protein from Arabidopsis thaliana thylakoid lumen.
To be Published
|
|
6UT9
 
 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[4] rotavirus strain BM5265 | | Descriptor: | Outer capsid protein VP4 | | Authors: | Xu, S, Stuckert, M, Burnside, R, McGinnis, K, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6VKX
 
 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[8] rotavirus strain BM13851 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Outer capsid protein VP4, TETRAETHYLENE GLYCOL | | Authors: | Xu, S, McGinnis, K.R, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | Descriptor: | M156R | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-07-05 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
2AZH
 
 | | Solution structure of iron-sulfur cluster assembly protein SUFU from Bacillus subtilis, with zinc bound at the active site. Northeast Structural Genomics Consortium target SR17 | | Descriptor: | SufU, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Aramini, J.M, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-10 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Zn-Ligated Fe-S Cluster Assembly Scaffold Protein SufU From Bacillus subtilis
To be published
|
|
2DSM
 
 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
6NIW
 
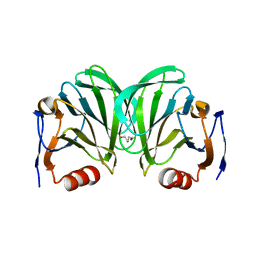 | | Crystal structure of P[6] rotavirus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease-sensitive outer capsid protein | | Authors: | Xu, S, Liu, Y, Lakamp, L, Ahmed, L, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
6OAI
 
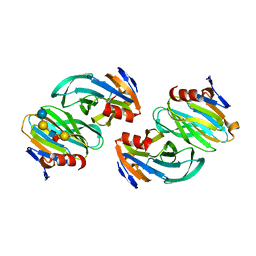 | | Crystal structure of P[6] rotavirus vp8* complexed with LNFPI | | Descriptor: | Protease-sensitive outer capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Xu, S, Liu, Y, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
2B3W
 
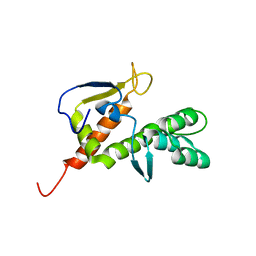 | | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24. | | Descriptor: | Hypothetical protein ybiA | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shih, L.Y, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-21 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24.
To be Published
|
|
2AKK
 
 | | Solution structure of phnA-like protein rp4479 from Rhodopseudomonas palustris | | Descriptor: | phnA-like protein | | Authors: | Wu, B, Yee, A, Ramelot, T.A, Semesi, A, Lemak, A, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2005-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of phnA-like protein rp4479 from Rhodopseudomonas palustris
To be Published
|
|
1D4U
 
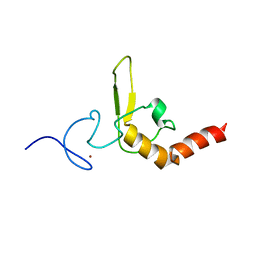 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
1ZG2
 
 | | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2. | | Descriptor: | Hypothetical UPF0213 protein BH0048 | | Authors: | Aramini, J.M, Swapna, G.V.T, Xiao, R, Ma, L, Shastry, R, Ciano, M, Acton, T.B, Liu, J, Rost, B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2.
To be Published
|
|
1EO1
 
 | |
1EIW
 
 | |
1XHS
 
 | | Solution NMR Structure of Protein ytfP from Escherichia coli. Northeast Structural Genomics Consortium Target ER111. | | Descriptor: | Hypothetical UPF0131 protein ytfP | | Authors: | Aramini, J.M, Huang, Y.J, Swapna, G.V.T, Paranji, R.K, Xiao, R, Shastry, R, Acton, T.B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Escherichia coli ytfP expands the structural coverage of the UPF0131 protein domain family.
Proteins, 68, 2007
|
|
1FGH
 
 | | COMPLEX WITH 4-HYDROXY-TRANS-ACONITATE | | Descriptor: | 4-HYDROXY-ACONITATE ION, ACONITASE, IRON/SULFUR CLUSTER | | Authors: | Lauble, H, Kennedy, M.C, Emptage, M.H, Beinert, H, Stout, C.D. | | Deposit date: | 1996-09-14 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The reaction of fluorocitrate with aconitase and the crystal structure of the enzyme-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1XPN
 
 | | NMR structure of P. aeruginosa protein PA1324: Northeast Structural Genomics Consortium target PaP1 | | Descriptor: | hypothetical protein PA1324 | | Authors: | Cort, J.R, Ni, S, Lockert, E.E, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170).
Protein Sci., 18, 2009
|
|
4FPW
 
 | | Crystal Structure of CalU16 from Micromonospora echinospora. Northeast Structural Genomics Consortium Target MiR12. | | Descriptor: | CalU16 | | Authors: | Seetharaman, J, Lew, S, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Phillips Jr, G.N, Kennedy, M.A, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
