4P09
 
 | | Crystal structure of HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31 | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
4P0A
 
 | | Crystal structure of HOIP PUB domain in complex with p97 PIM | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Transitional endoplasmic reticulum ATPase | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
4P0B
 
 | | Crystal structure of HOIP PUB domain in complex with OTULIN PIM | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin thioesterase otulin | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7005 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
4XKH
 
 | | CRYSTAL STRUCTURE OF THE AIRAPL TANDEM UIMS IN COMPLEX WITH A LYS48-LINKED TRI-UBIQUITIN | | Descriptor: | AN1-type zinc finger protein 2B, Polyubiquitin-C | | Authors: | Rahighi, S, Kawasaki, M, Stanhill, A, Wakatsuki, S. | | Deposit date: | 2015-01-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains.
Structure, 24, 2016
|
|
2IEY
 
 | | Crystal Structure of mouse Rab27b bound to GDP in hexagonal space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IF0
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IEZ
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4Z4K
 
 | |
4Z4M
 
 | |
7CFO
 
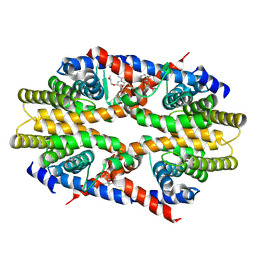 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
1UJJ
 
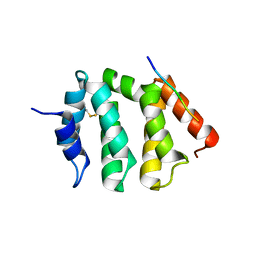 | | VHS domain of human GGA1 complexed with C-terminal peptide from BACE | | Descriptor: | ADP-ribosylation factor binding protein GGA1, C-terminal peptide from Beta-secretase | | Authors: | Shiba, T, Kametaka, S, Kawasaki, M, Shibata, M, Waguri, S, Uchiyama, Y, Wakatsuki, S. | | Deposit date: | 2003-08-05 | | Release date: | 2004-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Phosphoregulation of beta-Secretase Sorting Signal by the VHS Domain of GGA1
TRAFFIC, 5, 2004
|
|
1UJK
 
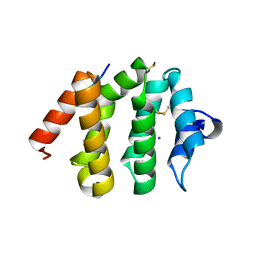 | | VHS domain of human GGA1 complexed with C-terminal phosphopeptide from BACE | | Descriptor: | ADP-ribosylation factor binding protein GGA1, C-terminal peptide from Beta-secretase, IODIDE ION | | Authors: | Shiba, T, Kametaka, S, Kawasaki, M, Shibata, M, Waguri, S, Uchiyama, Y, Wakatsuki, S. | | Deposit date: | 2003-08-05 | | Release date: | 2004-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Phosphoregulation of beta-Secretase Sorting Signal by the VHS Domain of GGA1
TRAFFIC, 5, 2004
|
|
2DWX
 
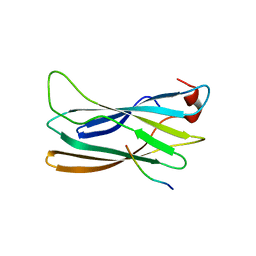 | | Co-crystal Structure Analysis of GGA1-GAE with the WNSF motif | | Descriptor: | ADP-ribosylation factor-binding protein GGA1, hinge peptide from ADP-ribosylation factor binding protein GGA1 | | Authors: | Inoue, M, Shiba, T, Yamada, Y, Ihara, K, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2006-08-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular Basis for Autoregulatory Interaction Between GAE Domain and Hinge Region of GGA1
Traffic, 8, 2007
|
|
2DWY
 
 | | Crystal Structure Analysis of GGA1-GAE | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA1 | | Authors: | Inoue, M, Shiba, T, Yamada, Y, Ihara, K, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2006-08-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Autoregulatory Interaction Between GAE Domain and Hinge Region of GGA1
Traffic, 8, 2007
|
|
7EH7
 
 | | Cryo-EM structure of the octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EH8
 
 | | Cryo-EM structure of the hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
1JWG
 
 | | VHS Domain of human GGA1 complexed with cation-independent M6PR C-terminal Peptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Cation-independent mannose-6-phosphate receptor, IODIDE ION | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1JWF
 
 | | Crystal Structure of human GGA1 VHS domain. | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1J2J
 
 | | Crystal structure of GGA1 GAT N-terminal region in complex with ARF1 GTP form | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor binding protein GGA1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-01-05 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
NAT.STRUCT.BIOL., 10, 2003
|
|
1IU1
 
 | | Crystal structure of human gamma1-adaptin ear domain | | Descriptor: | gamma1-adaptin | | Authors: | Nogi, T, Shiba, Y, Kawasaki, M, Shiba, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Takatsu, H, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2002-02-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain.
Nat.Struct.Biol., 9, 2002
|
|
7CN0
 
 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CN1
 
 | | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | | Descriptor: | POTASSIUM ION, potassium channel | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
3VHX
 
 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
7F3E
 
 | | Cryo-EM structure of the minimal protein-only RNase P from Aquifex aeolicus | | Descriptor: | RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Adachi, N, Kawasaki, M, Moriya, T, Numata, T, Senda, T, Kakuta, Y. | | Deposit date: | 2021-06-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Minimal protein-only RNase P structure reveals insights into tRNA precursor recognition and catalysis.
J.Biol.Chem., 297, 2021
|
|
3WXA
 
 | | X-ray crystal structural analysis of the complex between ALG-2 and Sec31A peptide | | Descriptor: | Programmed cell death protein 6, Protein transport protein Sec31A, ZINC ION | | Authors: | Takahashi, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Int J Mol Sci, 16, 2015
|
|
