2D7C
 
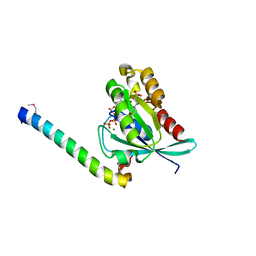 | | Crystal structure of human Rab11 in complex with FIP3 Rab-binding domain | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Shiba, T, Koga, H, Shin, H.W, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | 登録日 | 2005-11-16 | | 公開日 | 2006-09-26 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7WAF
 
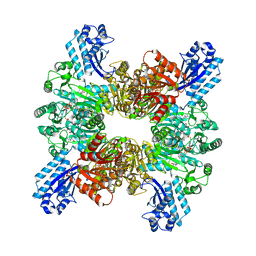 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | 分子名称: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | 著者 | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | 登録日 | 2021-12-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAE
 
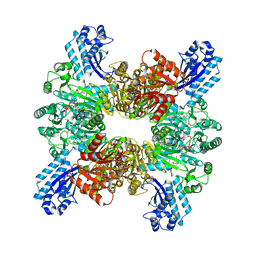 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS, 4x(beta-Asp-Arg), and aspartate | | 分子名称: | 4x(beta-Asp-Arg), ARGININE, ASPARTIC ACID, ... | | 著者 | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | 登録日 | 2021-12-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
2DX5
 
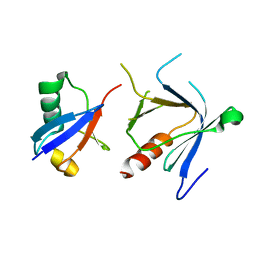 | | The complex structure between the mouse EAP45-GLUE domain and ubiquitin | | 分子名称: | Ubiquitin, Vacuolar protein sorting protein 36 | | 著者 | Hirano, S, Suzuki, N, Slagsvold, T, Kawasaki, M, Trambaiolo, D, Kato, R, Stenmark, H, Wakatsuki, S. | | 登録日 | 2006-08-24 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain
Nat.Struct.Mol.Biol., 13, 2006
|
|
7WLG
 
 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | 分子名称: | Alpha-xylosidase | | 著者 | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | 登録日 | 2022-01-13 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
2D3G
 
 | | Double sided ubiquitin binding of Hrs-UIM | | 分子名称: | ubiquitin, ubiquitin interacting motif from hepatocyte growth factor-regulated tyrosine kinase substrate | | 著者 | Hirano, S, Kawasaki, M, Kato, R, Wakatsuki, S. | | 登録日 | 2005-09-28 | | 公開日 | 2005-12-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting
Nat.Struct.Mol.Biol., 13, 2006
|
|
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | 分子名称: | BARIUM ION, TIP60 K67E mutant | | 著者 | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | 登録日 | 2022-04-24 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
6KNF
 
 | | CryoEM map and model of Nitrite Reductase at pH 6.2 | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase | | 著者 | Adachi, N, Yamaguchi, T, Moriya, T, Kawasaki, M, Koiwai, K, Shinoda, A, Yamada, Y, Yumoto, F, Kohzuma, T, Senda, T. | | 登録日 | 2019-08-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | 2.85 and 2.99 angstrom resolution structures of 110 kDa nitrite reductase determined by 200 kV cryogenic electron microscopy.
J.Struct.Biol., 213, 2021
|
|
6KNG
 
 | | CryoEM map and model of Nitrite Reductase at pH 8.1 | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase | | 著者 | Adachi, N, Yamaguchi, T, Moriya, T, Kawasaki, M, Koiwai, K, Shinoda, A, Yamada, Y, Yumoto, F, Kohzuma, T, Senda, T. | | 登録日 | 2019-08-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | 2.85 and 2.99 angstrom resolution structures of 110 kDa nitrite reductase determined by 200 kV cryogenic electron microscopy.
J.Struct.Biol., 213, 2021
|
|
2ZND
 
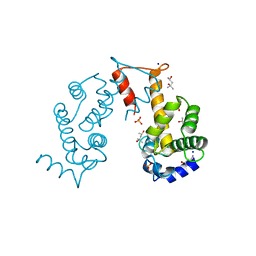 | | Crystal structure of Ca2+-free form of des3-20ALG-2 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHATE ION, Programmed cell death protein 6, ... | | 著者 | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | 登録日 | 2008-04-22 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZAO
 
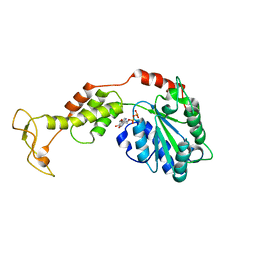 | | Crystal structure of mouse SKD1/VPS4B ADP-form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Vacuolar protein sorting-associating protein 4B | | 著者 | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2007-10-08 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZNE
 
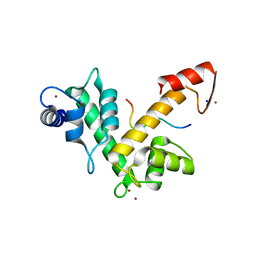 | | Crystal structure of Zn2+-bound form of des3-23ALG-2 complexed with Alix ABS peptide | | 分子名称: | 16-meric peptide from Programmed cell death 6-interacting protein, Programmed cell death protein 6, SODIUM ION, ... | | 著者 | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | 登録日 | 2008-04-22 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZAN
 
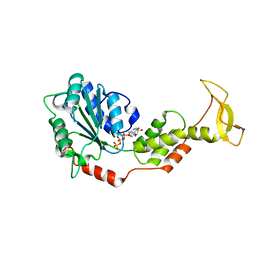 | | Crystal structure of mouse SKD1/VPS4B ATP-form | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Vacuolar protein sorting-associating protein 4B | | 著者 | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2007-10-08 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZN8
 
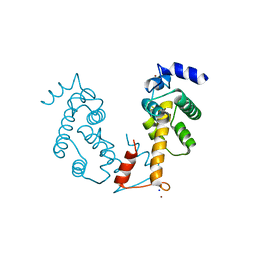 | | Crystal structure of Zn2+-bound form of ALG-2 | | 分子名称: | Programmed cell death protein 6, SODIUM ION, ZINC ION | | 著者 | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | 登録日 | 2008-04-22 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZAM
 
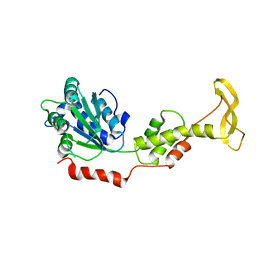 | | Crystal structure of mouse SKD1/VPS4B apo-form | | 分子名称: | Vacuolar protein sorting-associating protein 4B | | 著者 | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2007-10-08 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZN9
 
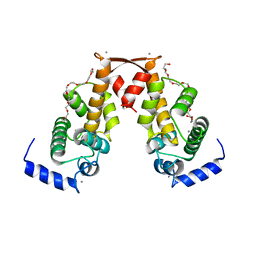 | | Crystal structure of Ca2+-bound form of des3-20ALG-2 | | 分子名称: | CALCIUM ION, DODECAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | 著者 | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | 登録日 | 2008-04-22 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
3VTV
 
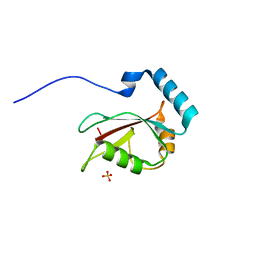 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | 分子名称: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | 著者 | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | 登録日 | 2012-06-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3WAL
 
 | | Crystal structure of human LC3A_2-121 | | 分子名称: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2013-05-06 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAO
 
 | | Crystal structure of Atg13 LIR-fused human LC3B_2-119 | | 分子名称: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3B | | 著者 | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2013-05-06 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAN
 
 | | Crystal structure of Atg13 LIR-fused human LC3A_2-121 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2013-05-06 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAM
 
 | | Crystal structure of human LC3C_8-125 | | 分子名称: | CITRIC ACID, Microtubule-associated proteins 1A/1B light chain 3C | | 著者 | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2013-05-06 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAP
 
 | | Crystal structure of Atg13 LIR-fused human LC3C_8-125 | | 分子名称: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3C | | 著者 | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2013-05-06 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WZ2
 
 | | Crystal structure of Pyrococcus furiosus PbaA, an archaeal homolog of proteasome-assembly chaperone | | 分子名称: | Uncharacterized protein | | 著者 | Sikdar, A, Satoh, T, Kawasaki, M, Kato, K. | | 登録日 | 2014-09-18 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of archaeal homolog of proteasome-assembly chaperone PbaA
Biochem.Biophys.Res.Commun., 453, 2014
|
|
2ZVN
 
 | | NEMO CoZi domain incomplex with diubiquitin in P212121 space group | | 分子名称: | NF-kappa-B essential modulator, UBC protein | | 著者 | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | 登録日 | 2008-11-12 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
3AAK
 
 | | Crystal structure of Zn2+-bound form of des3-20ALG-2F122A | | 分子名称: | Programmed cell death protein 6, ZINC ION | | 著者 | Inuzuka, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | 登録日 | 2009-11-19 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
