2GVE
 
 | | Time-of-Flight Neutron Diffraction Structure of D-Xylose Isomerase | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Li, X, Carrell, H.L, Hanson, B.L, Langan, P, Coates, L, Schoenborn, B.P, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-05-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GLK
 
 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7B0B
 
 | | Fab HbnC3t1p1_C6 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, IGK@ protein, ... | | Authors: | Borenstein-Katz, A, Diskin, R. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants.
Immunity, 56, 2023
|
|
8BF0
 
 | |
6GC2
 
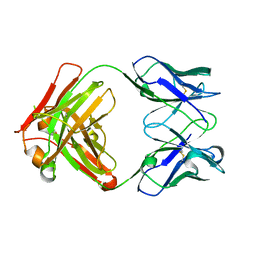 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
6XUD
 
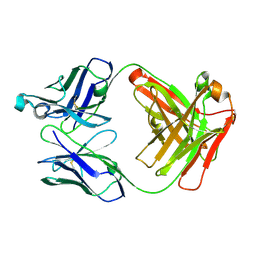 | | Apo Ab 1116NS19.9 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUL
 
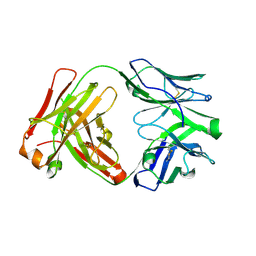 | | Apo Ab 5b1 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XTG
 
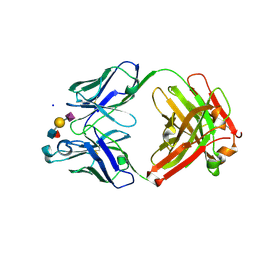 | | Ab 1116NS19.9 bound to CA19-9 | | Descriptor: | Heavy chain, Light chain, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUK
 
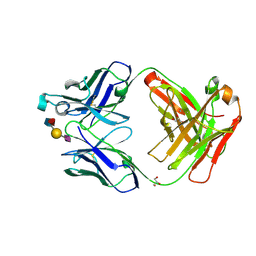 | | AbLIFT design 15 of Ab 1116NS19.9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUN
 
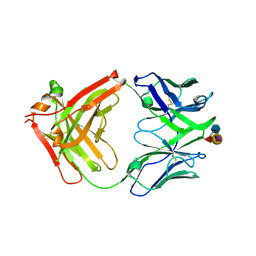 | | Ab 5b1 bound to CA19-9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
4ISV
 
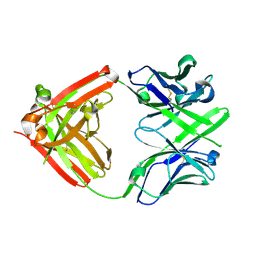 | | Crystal structure of the Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC FOR POLY-GLUTAMINE | | Descriptor: | 1C2 FAB HEAVY CHAIN, 1C2 FAB LIGHT CHAIN | | Authors: | Klein, F.A.C, Zeder-Lutz, G, Cousido-Siah, A, Mitschler, A, Katz, A, Eberling, P, Mandel, J.L, Podjarny, A, Trottier, Y. | | Deposit date: | 2013-01-17 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10.
Hum.Mol.Genet., 22, 2013
|
|
4JJ5
 
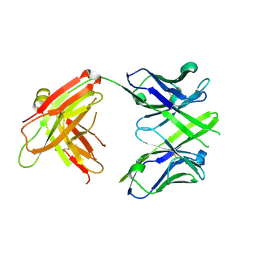 | | CRYSTAL STRUCTURE OF THE Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC for POLY-GLUTAMINE | | Descriptor: | 1C2 FAB HEAVY CHAIN, 1C2 FAB LIGHT CHAIN | | Authors: | Klein, F.A.C, Zeder-Lutz, G, Cousido-Siah, A, Mitschler, A, Katz, A, Eberling, P, Mandel, J.L, Podjarny, A, Trottier, Y. | | Deposit date: | 2013-03-07 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10.
Hum.Mol.Genet., 22, 2013
|
|
1S1G
 
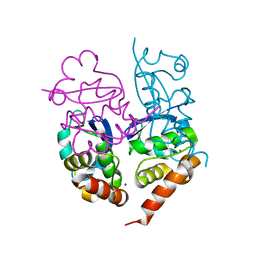 | | Crystal Structure of Kv4.3 T1 Domain | | Descriptor: | Potassium voltage-gated channel subfamily D member 3, ZINC ION | | Authors: | Scannevin, R.H, Wang, K.W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.X, Xu, Z.B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
2GUB
 
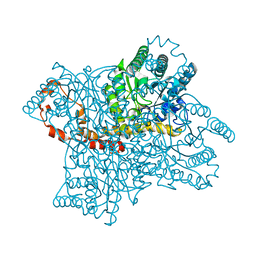 | |
1Q4G
 
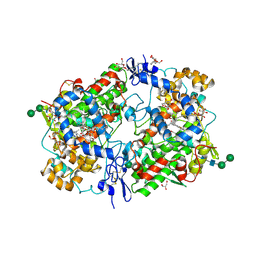 | | 2.0 Angstrom Crystal Structure of Ovine Prostaglandin H2 Synthase-1, in complex with alpha-methyl-4-biphenylacetic acid | | Descriptor: | 2-(1,1'-BIPHENYL-4-YL)PROPANOIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Gupta, K, Selinksy, B.S, Kaub, C.J, Katz, A.K, Loll, P.J. | | Deposit date: | 2003-08-03 | | Release date: | 2004-01-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A Resolution Crystal Structure of Prostaglandin H(2) Synthase-1: Structural Insights into an Unusual Peroxidase
J.Mol.Biol., 335, 2004
|
|
8PF0
 
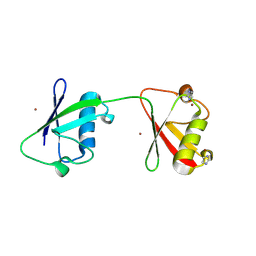 | | Diubiquitin-derived artificial binding protein (Affilin) variant Af1 specific to oncofetal fibronectin | | Descriptor: | Affilin variant Af1 (77405), COPPER (II) ION, IMIDAZOLE | | Authors: | Parthier, C, Katzschmann, A, Fiedler, E, Haupts, U, Reimann, A. | | Deposit date: | 2023-06-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ubiquitin-derived artificial binding proteins targeting oncofetal fibronectin reveal scaffold plasticity by beta-strand slippage.
Commun Biol, 7, 2024
|
|
8PEQ
 
 | | Complex of diubiquitin-derived artificial binding protein (Affilin) variant Af2 with its target oncofetal fibronectin (fragment 7B8) | | Descriptor: | Affilin variant Af2, Fibronectin, SULFATE ION | | Authors: | Parthier, C, Katzschmann, A, Fiedler, E, Haupts, U, Reimann, A. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Ubiquitin-derived artificial binding proteins targeting oncofetal fibronectin reveal scaffold plasticity by beta-strand slippage.
Commun Biol, 7, 2024
|
|
8FWY
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to the dead-end complex xanthine and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, XANTHINE | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX3
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-GP, showing the structure of the complete active site in its open conformation | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX1
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (R)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX0
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (S)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX2
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-HP | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FWZ
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Hydroxypropyl-Lin-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, [(2R)-2-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}propyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
