1XJ9
 
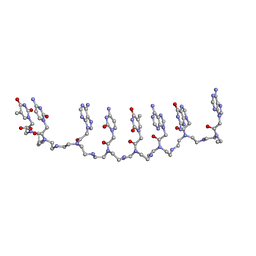 | | Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network | | 分子名称: | peptide nucleic acid, (H-P(*GPN*TPN*APN*GPN*APN*TPN*CPN*APN*CPN*TPN)-LYS-NH2) | | 著者 | Petersson, B, Nielsen, B.B, Rasmussen, H, Larsen, I.K, Gajhede, M, Nielsen, P.E, Kastrup, J.S. | | 登録日 | 2004-09-23 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of a Partly Self-Complementary Peptide Nucleic Acid (PNA) Oligomer Showing a Duplex-Triplex Network
J.Am.Chem.Soc., 127, 2005
|
|
6GL4
 
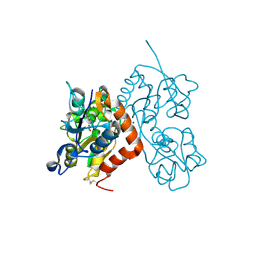 | | Structure of GluA2o ligand-binding domain (S1S2J) in complex with glutamate and sodium bromide at 1.95 A resolution | | 分子名称: | ACETATE ION, BROMIDE ION, GLUTAMIC ACID, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-05-22 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.948 Å) | | 主引用文献 | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
2WIM
 
 | | Crystal structure of NCAM2 IG1-3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAL CELL ADHESION MOLECULE 2 | | 著者 | Kulahin, N, Kristensen, O, Rasmussen, K, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | 登録日 | 2009-05-13 | | 公開日 | 2010-08-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural model and trans-interaction of the entire ectodomain of the olfactory cell adhesion molecule.
Structure, 19, 2011
|
|
3C9N
 
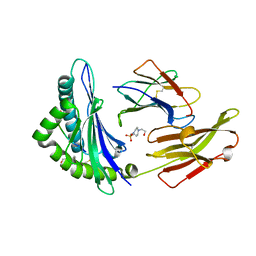 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | 登録日 | 2008-02-18 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
4NWD
 
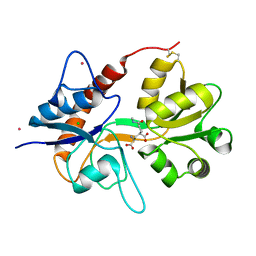 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | 分子名称: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | 分子名称: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.012 Å) | | 主引用文献 | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
6GIV
 
 | | Structure of GluA2-N775S ligand-binding domain (S1S2J) in complex with glutamate and Rubidium Bromide at 1.75 A resolution | | 分子名称: | BROMIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-05-15 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
5O9A
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504Y-N775S) in complex with glutamate and BPAM121 at 1.78 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | 著者 | Laulumaa, S, Rovinskaja, K, Frydenvang, K.A, Kastrup, J.S. | | 登録日 | 2017-06-16 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
2VAJ
 
 | | Crystal structure of NCAM2 Ig1 (I4122 cell unit) | | 分子名称: | NEURAL CELL ADHESION MOLECULE 2 | | 著者 | Kulahin, N, Rasmussen, K.K, Kristensen, O, Kastrup, J.S, Navarro-Poulsen, J.-C, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | 登録日 | 2007-08-31 | | 公開日 | 2008-08-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Crystal Structure of the Ig1 Domain of the Neural Cell Adhesion Molecule Ncam2 Displays Domain Swapping.
J.Mol.Biol., 382, 2008
|
|
2WKY
 
 | | Crystal structure of the ligand-binding core of GluR5 in complex with the agonist 4-AHCP | | 分子名称: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | 著者 | Naur, P, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-06-18 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Glutamate Receptor Glur5 Agonist (S)-2-Amino-3-(3-Hydroxy-7,8-Dihydro-6H-Cyclohepta[D]Isoxazol-4-Yl)Propionic Acid and the 8-Methyl Analogue: Synthesis, Molecular Pharmacology, and Biostructural Characterization
J.Med.Chem., 52, 2009
|
|
1R7A
 
 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | 著者 | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | 登録日 | 2003-10-21 | | 公開日 | 2004-02-10 | | 最終更新日 | 2014-11-19 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
6HCC
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 1.6 A RESOLUTION. | | 分子名称: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), ACETATE ION, CHLORIDE ION, ... | | 著者 | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-08-14 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.617 Å) | | 主引用文献 | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HCB
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.9 A RESOLUTION. | | 分子名称: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Laulumaa, S, Masternak, M, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-08-14 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HCA
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S1J) IN COMPLEX WITH POSITIVE ALLOSTERIC MODULATOR TDPAM02 AT 1.8 A RESOLUTION | | 分子名称: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-08-14 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.882 Å) | | 主引用文献 | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HCH
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.6 A RESOLUTION. | | 分子名称: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), ACETATE ION, CHLORIDE ION, ... | | 著者 | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-08-15 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HC9
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 2.4 A RESOLUTION. | | 分子名称: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-08-14 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
1EPF
 
 | | CRYSTAL STRUCTURE OF THE TWO N-TERMINAL IMMUNOGLOBULIN DOMAINS OF THE NEURAL CELL ADHESION MOLECULE (NCAM) | | 分子名称: | CALCIUM ION, PROTEIN (NEURAL CELL ADHESION MOLECULE) | | 著者 | Kasper, C, Rasmussen, H, Kastrup, J.S, Ikemizu, S, Jones, E.Y, Berezin, V, Bock, E, Larsen, I.K. | | 登録日 | 2000-03-29 | | 公開日 | 2000-10-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis of cell-cell adhesion by NCAM.
Nat.Struct.Biol., 7, 2000
|
|
3H6W
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution | | 分子名称: | (3R)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6U
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution | | 分子名称: | (3S)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CITRATE ANION, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3S2V
 
 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | 分子名称: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2011-05-17 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | 分子名称: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | 著者 | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | 登録日 | 2002-07-09 | | 公開日 | 2002-09-18 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5B
 
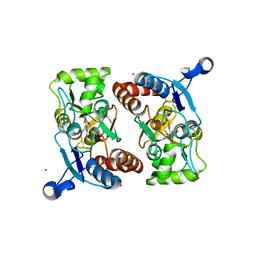 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | 著者 | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | 登録日 | 2002-07-09 | | 公開日 | 2002-09-18 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | 分子名称: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | 著者 | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | 登録日 | 2002-07-09 | | 公開日 | 2002-09-18 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
3H6V
 
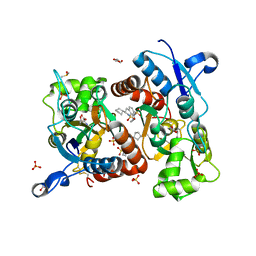 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | 分子名称: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
4NZB
 
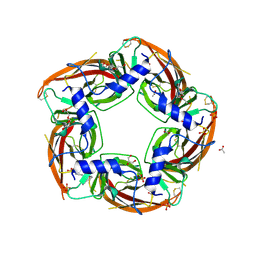 | | NS9283 bound to Ls-AChBP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[3-(pyridin-3-yl)-1,2,4-oxadiazol-5-yl]benzonitrile, ACETATE ION, ... | | 著者 | Olsen, J.A, Kastrup, J.S, Gajhede, M. | | 登録日 | 2013-12-11 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural and functional studies of the modulator NS9283 reveal agonist-like mechanism of action at alpha 4 beta 2 nicotinic acetylcholine receptors.
J.Biol.Chem., 289, 2014
|
|
