3B97
 
 | | Crystal Structure of human Enolase 1 | | Descriptor: | Alpha-enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Kang, H.J, Jung, S.K, Kim, S.J, Chung, S.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3GLD
 
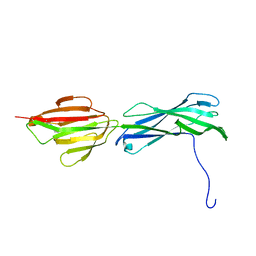 | |
3HTL
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly with stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, Putative surface-anchored fimbrial subunit, SODIUM ION | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-11 | | Release date: | 2009-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HR6
 
 | | Structure of the Corynebacterium diphtheriae major pilin SpaA points to a modular pilus assembly stabilizing isopeptide bonds | | Descriptor: | CALCIUM ION, IODIDE ION, Putative surface-anchored fimbrial subunit | | Authors: | Kang, H.J, Paterson, N.G, Gaspar, A.H, Ton-That, H, Baker, E.N. | | Deposit date: | 2009-06-08 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GLE
 
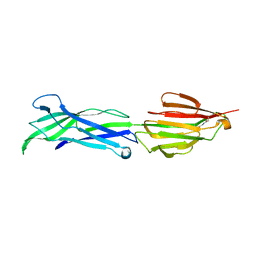 | |
3PSQ
 
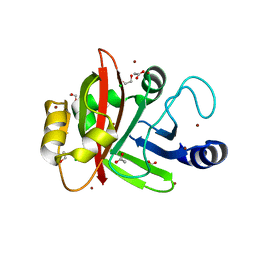 | | Crystal structure of Spy0129, a Streptococcus pyogenes class B sortase involved in pilus biogenesis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kang, H.J, Baker, E.N. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of Spy0129, a Streptococcus pyogenes class B sortase involved in pilus assembly
Plos One, 6, 2011
|
|
3B2M
 
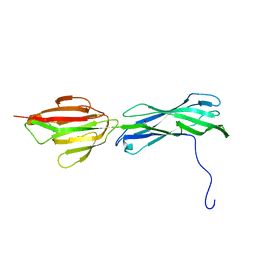 | |
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZL9
 
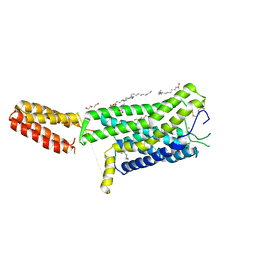 | | Crystal structure of human GPCR Niacin receptor (HCA2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, FiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZLY
 
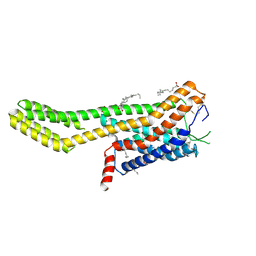 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
4DCO
 
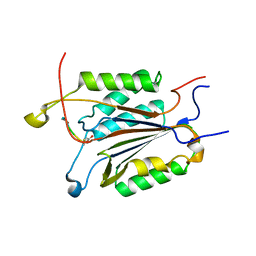 | | Crystal Structure of caspase 3, L168Y mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4DCJ
 
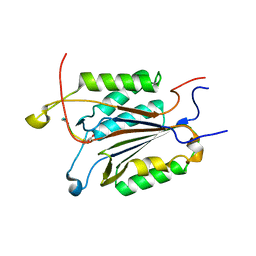 | | Crystal structure of caspase 3, L168D mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4HQR
 
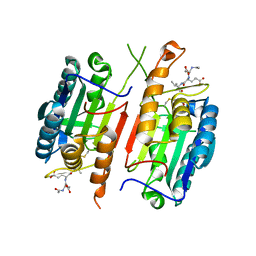 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Ac-Asp-Glu-Val-Asp-Aldehyde, Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-26 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HSS
 
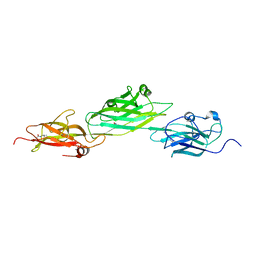 | |
4HSQ
 
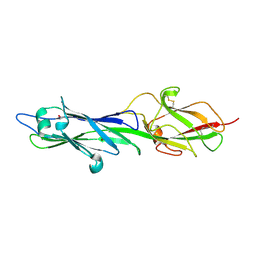 | |
4HQ0
 
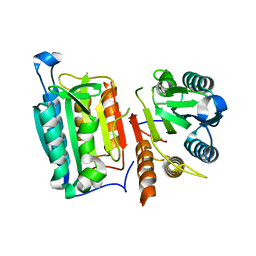 | | Crystal Structure of mutant form of Caspase-7 | | Descriptor: | Caspase-7 | | Authors: | Lee, Y, Kang, H.J, Bae, K.-H, Kim, S.J, Chung, S.J. | | Deposit date: | 2012-10-25 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural asymmetry of procaspase-7 bound to a specific inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4DCP
 
 | | Crystal Structure of caspase 3, L168F mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4K8W
 
 | | An arm-swapped dimer of the S. pyogenes pilin specific assembly factor SipA | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, LepA | | Authors: | Young, P.G, Kang, H.J, Baker, E.N. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An arm-swapped dimer of the Streptococcus pyogenes pilin specific assembly factor SipA.
J.Struct.Biol., 183, 2013
|
|
4YB9
 
 | | Crystal structure of the Bovine Fructose transporter GLUT5 in an open inward-facing conformation | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5 | | Authors: | Verdon, G, Kang, H.J, Iwata, S, Drew, D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5.
Nature, 526, 2015
|
|
