3DUZ
 
 | | Crystal structure of the postfusion form of baculovirus fusion protein GP64 | | Descriptor: | MERCURY (II) ION, Major envelope glycoprotein | | Authors: | Kadlec, J, Loureiro, S, Abrescia, N.G.A, Jones, I.M, Stuart, D.I. | | Deposit date: | 2008-07-18 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2Y0N
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MSL3 | | Descriptor: | MALE-SPECIFIC LETHAL 1 HOMOLOG, MALE-SPECIFIC LETHAL 3 HOMOLOG | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-04 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2Y0M
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MOF | | Descriptor: | ACETYL COENZYME *A, MALE-SPECIFIC LETHAL 1 HOMOLOG, PROBABLE HISTONE ACETYLTRANSFERASE MYST1, ... | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2IYK
 
 | | Crystal structure of the UPF2-interacting domain of nonsense mediated mRNA decay factor UPF1 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, ZINC ION | | Authors: | Kadlec, J, Guilligay, D, Ravelli, R.B, Cusack, S. | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Upf2-Interacting Domain of Nonsense-Mediated Mrna Decay Factor Upf1.
RNA, 12, 2006
|
|
1UW4
 
 | |
7QWV
 
 | | Crystal structure of the REC114-TOPOVIBL complex. | | Descriptor: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | Authors: | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
7QUU
 
 | | Red1-Iss10 complex | | Descriptor: | NURS complex subunit red1, Uncharacterized protein C7D4.14c | | Authors: | Mackereth, C.D, Kadlec, J, Laroussi, H. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
7QY5
 
 | | Crystal structure of the S.pombe Ars2-Red1 complex. | | Descriptor: | NURS complex subunit pir2, RNA elimination defective protein Red1, ZINC ION | | Authors: | Foucher, A.E, Kadlec, J. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
4UOI
 
 | | Unexpected structure for the N-terminal domain of Hepatitis C virus envelope glycoprotein E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GENOME POLYPROTEIN | | Authors: | El Omari, K, Iourin, O, Kadlec, J, Harlos, K, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2014-06-04 | | Release date: | 2014-08-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Unexpected Structure for the N-Terminal Domain of Hepatitis C Virus Envelope Glycoprotein E1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5O8M
 
 | |
8BY6
 
 | | Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
6HFG
 
 | |
1L4A
 
 | | X-RAY STRUCTURE OF THE NEURONAL COMPLEXIN/SNARE COMPLEX FROM THE SQUID LOLIGO PEALEI | | Descriptor: | S-SNAP25 fusion protein, S-SYNTAXIN, SYNAPHIN A, ... | | Authors: | Bracher, A, Kadlec, J, Betz, H, Weissenhorn, W. | | Deposit date: | 2002-03-04 | | Release date: | 2002-07-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | X-ray structure of a neuronal complexin-SNARE complex from squid.
J.Biol.Chem., 277, 2002
|
|
8PNT
 
 | | Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8PMP
 
 | | Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
2WJV
 
 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, REGULATOR OF NONSENSE TRANSCRIPTS 2, SULFATE ION, ... | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
2WJY
 
 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 Orthorhombic form | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, SULFATE ION, ZINC ION | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
4CY2
 
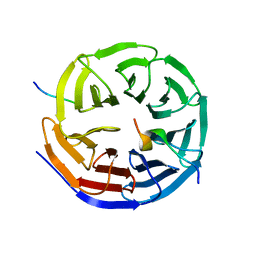 | | Crystal structure of the KANSL1-WDR5-KANSL2 complex. | | Descriptor: | KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, KAT8 REGULATORY NSL COMPLEX SUBUNIT 2, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CY3
 
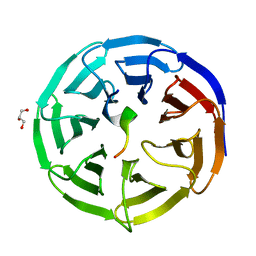 | | Crystal structure of the NSL1-WDS complex. | | Descriptor: | CG4699, ISOFORM D, GLYCEROL, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CY5
 
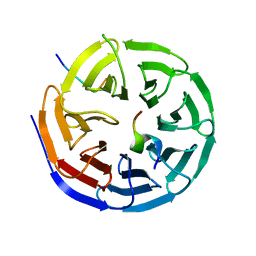 | | Crystal structure of the NSL1-WDS-NSL2 complex. | | Descriptor: | CG4699, ISOFORM D, DIM GAMMA-TUBULIN 1, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CY1
 
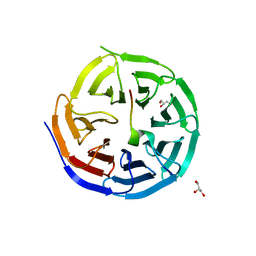 | | Crystal structure of the KANSL1-WDR5 complex. | | Descriptor: | GLYCEROL, KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4D25
 
 | | Crystal structure of the Bombyx mori Vasa helicase (E339Q) in complex with RNA and AMPPNP | | Descriptor: | 5'-R(*UP*GP*AP*CP*AP*UP)-3', BMVLG PROTEIN, GLYCEROL, ... | | Authors: | Spinelli, P, Pillai, R.S, Kadlec, J, Cusack, S. | | Deposit date: | 2014-05-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RNA Clamping by Vasa Assembles a Pirna Amplifier Complex on Transposon Transcripts.
Cell(Cambridge,Mass.), 157, 2014
|
|
4D26
 
 | | Crystal structure of the Bombyx mori Vasa helicase (E339Q) in complex with RNA,ADP and Pi | | Descriptor: | 5'-R(*UP*GP*AP*CP*AP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, BMVLG PROTEIN, ... | | Authors: | Spinelli, P, Pillai, R.S, Kadlec, J, Cusack, S. | | Deposit date: | 2014-05-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RNA Clamping by Vasa Assembles a Pirna Amplifier Complex on Transposon Transcripts.
Cell(Cambridge,Mass.), 157, 2014
|
|
4B7Y
 
 | | Crystal structure of the MSL1-MSL2 complex | | Descriptor: | MALE-SPECIFIC LETHAL 1 HOMOLOG, MALE-SPECIFIC LETHAL 2 HOMOLOG, ZINC ION | | Authors: | Hallacli, E, Lipp, M, Georgiev, P, Spielman, C, Cusack, S, Akhtar, A, Kadlec, J. | | Deposit date: | 2012-08-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Msl1-Mediated Dimerization of the Dosage Compensation Complex is Essential for Male X-Chromosome Regulation in Drosophila.
Mol.Cell, 48, 2012
|
|
4B86
 
 | | Crystal structure of the MSL1-MSL2 complex (3.5A) | | Descriptor: | MALE-SPECIFIC LETHAL 1 HOMOLOG, MALE-SPECIFIC LETHAL 2 HOMOLOG, ZINC ION | | Authors: | Hallacli, E, Lipp, M, Georgiev, P, Spielman, C, Cusack, S, Akhtar, A, Kadlec, J. | | Deposit date: | 2012-08-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Msl1-Mediated Dimerization of the Dosage Compensation Complex is Essential for Male X-Chromosome Regulation in Drosophila.
Mol.Cell, 48, 2012
|
|
