3OUH
 
 | | PHD2-R127 with JNJ41536014 | | Descriptor: | 1-(5-chloro-6-fluoro-1H-benzimidazol-2-yl)-1H-pyrazole-4-carboxylic acid, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Kim, H, Clark, R. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
5Y7X
 
 | | Human Peroxisome proliferator-activated receptor (PPAR) delta in complexed with a potent and selective agonist | | Descriptor: | 2-[2-methyl-4-[[4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-selenazol-5-yl]methylsulfanyl]phenoxy]ethanoic acid, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Kim, H.L, Chin, J.W, Cho, S.J, Song, J.Y, Yoon, H.S, Bae, J.H. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Design, synthesis, and the X-ray co-crystal structure of Highly Potent, Selective, and Orally Bioavailable, Novel Peroxisome Proliferator-Activated Receptor delta Agonists
To Be Published
|
|
4KBZ
 
 | |
3V9B
 
 | |
1KU0
 
 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
5X9Q
 
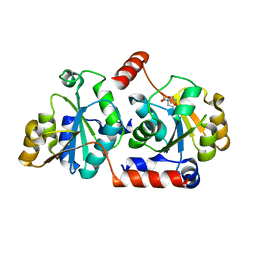 | | Crystal structure of HldC from Burkholderia pseudomallei | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of D-glycero-Beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei.
Proteins, 86, 2018
|
|
5XHW
 
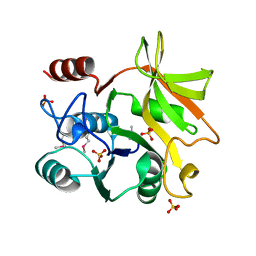 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
6XAV
 
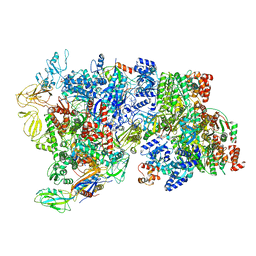 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
6XAS
 
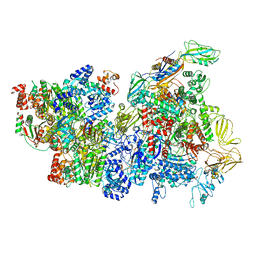 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
1UAJ
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAM
 
 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
3X2X
 
 | | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima | | Descriptor: | MANGANESE (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Crystal structure of metallo-beta-lactamase H48A from Thermotoga maritima
To be Published
|
|
6OIC
 
 | | Crystal structure of human Sulfide Quinone Oxidoreductase in complex with coenzyme Q (sulfite soaked) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, Sulfide:quinone oxidoreductase, ... | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-09 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
3X2Z
 
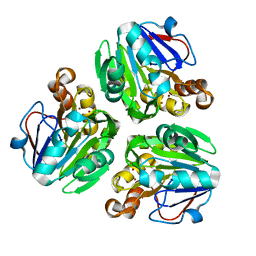 | | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima | | Descriptor: | NICKEL (II) ION, UPF0173 metal-dependent hydrolase TM_1162 | | Authors: | Choi, H.J, Kim, H.J, Matsuura, A, Mikami, B, Yoon, H.J, Lee, H.H. | | Deposit date: | 2015-01-07 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of metallo-beta-lactamase in complex with nickel from Thermotoga maritima
To be Published
|
|
6XKT
 
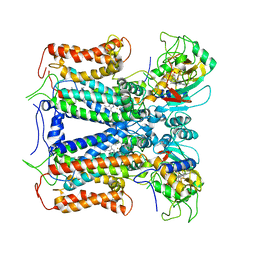 | | R. capsulatus cyt bc1 with both FeS proteins in c position (CIII2 c-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XI0
 
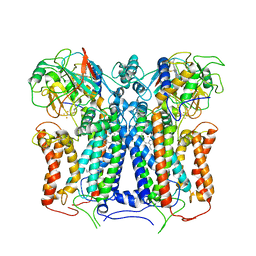 | | R. capsulatus cyt bc1 (CIII2) at 3.3A | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKU
 
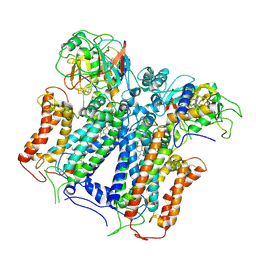 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKZ
 
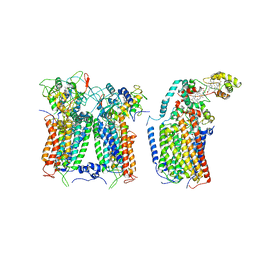 | | R. capsulatus CIII2CIV tripartite super-complex, conformation B (SC-1B) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
5XD8
 
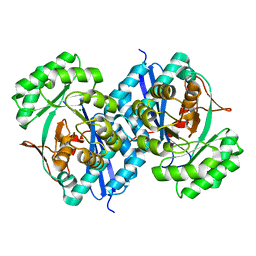 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
2DS8
 
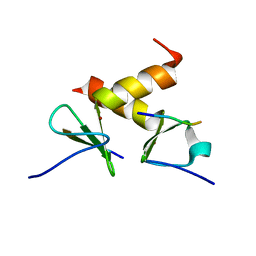 | | Structure of the ZBD-XB complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
5XD7
 
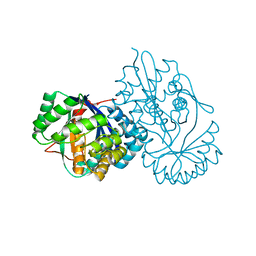 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, ACETIC ACID, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5XD0
 
 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | Authors: | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
8I9J
 
 | | The PKR and E3L complex | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase, RNA-binding protein E3 | | Authors: | Han, C.W, Kim, H.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.39 Å) | | Cite: | Structural study of novel vaccinia virus E3L and dsRNA-dependent protein kinase complex.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
6LHU
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHS
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
