5F51
 
 | | Structure of B. abortus WrbA-related protein A (apo) | | 分子名称: | NAD(P)H dehydrogenase (quinone), SULFATE ION | | 著者 | Herrou, J, Czyz, D, Willett, J.W, Kim, H.S, Kim, Y, Crosson, S. | | 登録日 | 2015-12-03 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | WrpA Is an Atypical Flavodoxin Family Protein under Regulatory Control of the Brucella abortus General Stress Response System.
J.Bacteriol., 198, 2016
|
|
7V8U
 
 | | Crystal structure of PsEst3 wild-type | | 分子名称: | Esterase, NITROBENZENE, SULFATE ION | | 著者 | Son, J, Kim, H, Kim, H.W. | | 登録日 | 2021-08-23 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Iucrj, 10, 2023
|
|
7V8X
 
 | |
7V8V
 
 | |
7V8W
 
 | | Crystal structure of PsEst3 S128A variant complexed with malonate | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, MALONIC ACID, ... | | 著者 | Son, J, Kim, H, Kim, H.W. | | 登録日 | 2021-08-23 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Iucrj, 10, 2023
|
|
5Y86
 
 | | Crystal structure of kinase | | 分子名称: | 1,2-ETHANEDIOL, 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 3, ... | | 著者 | Kim, K.L, Cha, J.S, Cho, Y.S, Kim, H.Y, Chang, N.P, Cho, H.S. | | 登録日 | 2017-08-18 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
J. Mol. Biol., 430, 2018
|
|
4G4V
 
 | |
4G4Y
 
 | |
7WWP
 
 | | Crystal structure of human Npl4 | | 分子名称: | Nuclear protein localization protein 4 homolog, ZINC ION | | 著者 | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | 登録日 | 2022-02-14 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
4G4X
 
 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | 分子名称: | D-ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | 著者 | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | 登録日 | 2012-07-16 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
To be Published
|
|
4G88
 
 | | Crystal structure of OmpA peptidoglycan-binding domain from Acinetobacter baumannii | | 分子名称: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein Omp38, S,R MESO-TARTARIC ACID | | 著者 | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | 登録日 | 2012-07-22 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
to be published
|
|
4G4Z
 
 | |
7WWQ
 
 | | Crystal structure of human Ufd1-Npl4 complex | | 分子名称: | Nuclear protein localization protein 4 homolog, Ubiquitin recognition factor in ER-associated degradation protein 1 | | 著者 | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | 登録日 | 2022-02-14 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
3INA
 
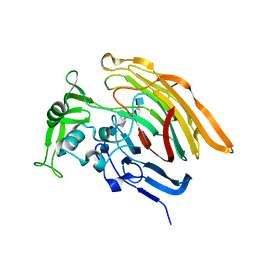 | | Crystal structure of heparin lyase I H151A mutant complexed with a dodecasaccharide heparin | | 分子名称: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, Heparin lyase I | | 著者 | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | 登録日 | 2009-08-12 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
4G4W
 
 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | 分子名称: | ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | 著者 | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | 登録日 | 2012-07-16 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
to be published
|
|
3IN9
 
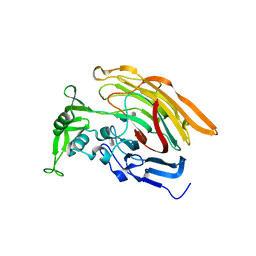 | | Crystal structure of heparin lyase I complexed with disaccharide heparin | | 分子名称: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparin lyase I | | 著者 | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | 登録日 | 2009-08-12 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
5XTF
 
 | |
1Q1Y
 
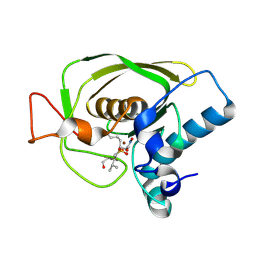 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | 分子名称: | ACTINONIN, Peptide deformylase, ZINC ION | | 著者 | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | 登録日 | 2003-07-23 | | 公開日 | 2004-07-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | 分子名称: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Han, A, Kim, H.M. | | 登録日 | 2022-12-19 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
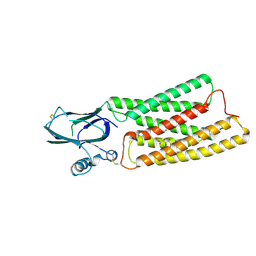 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | 著者 | Han, A, Kim, H.M. | | 登録日 | 2022-12-21 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
5FA9
 
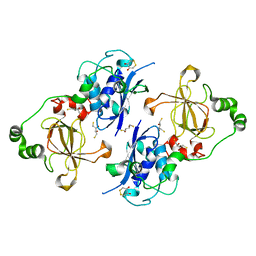 | | Bifunctional Methionine Sulfoxide Reductase AB (MsrAB) from Treponema denticola | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptide methionine sulfoxide reductase MsrA | | 著者 | Han, A, Son, J, Kim, H.-Y, Hwang, K.Y. | | 登録日 | 2015-12-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Essential Role of the Linker Region in the Higher Catalytic Efficiency of a Bifunctional MsrA-MsrB Fusion Protein
Biochemistry, 55, 2016
|
|
8YBE
 
 | |
4M6R
 
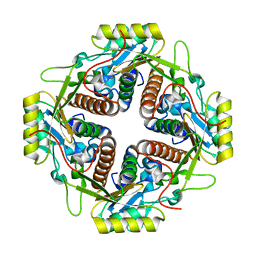 | | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme | | 分子名称: | Methylthioribulose-1-phosphate dehydratase, ZINC ION | | 著者 | Kang, W, Hong, S.H, Lee, H.M, Kim, N.Y, Lim, Y.C, Le, L.T.M, Lim, B, Kim, H.C, Kim, T.Y, Ashida, H, Yokota, A, Hah, S.S, Chun, K.H, Jung, Y.K, Yang, J.K. | | 登録日 | 2013-08-10 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4JJT
 
 | | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv | | 分子名称: | ACETATE ION, Enoyl-CoA hydratase, GLYCEROL | | 著者 | Tan, K, Holowicki, J, Endres, M, Kim, C.-Y, Kim, H, Hung, L.-W, Terwilliger, T.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2013-03-08 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.496 Å) | | 主引用文献 | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
5G1D
 
 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | 分子名称: | SYNDECAN-4, SYNTENIN-1 | | 著者 | Lee, I, Kim, H, Yun, J.H, Lee, W. | | 登録日 | 2016-03-25 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
