3GS3
 
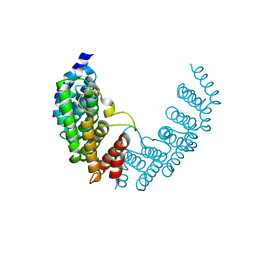 | |
2O83
 
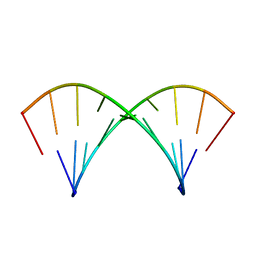 | |
2LX1
 
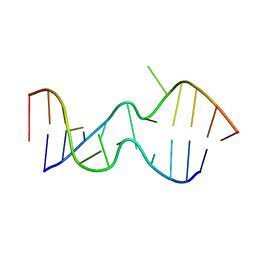 | |
4YND
 
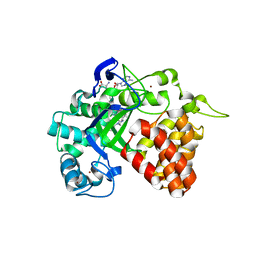 | | The Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2 | | 分子名称: | N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[(2R)-2-hydroxy-2-(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | 著者 | Sweis, R.F, Wang, Z, Algire, M, Arrowsmith, C.H, Brown, P.J, Chiang, G.C, Guo, J, Jakob, C.G, Kennedy, S, Li, F, Soni, N.B, Vedadi, M, Pappano, W.N. | | 登録日 | 2015-03-09 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2.
Acs Med.Chem.Lett., 6, 2015
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | 分子名称: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | 著者 | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | 登録日 | 2014-12-15 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6AU2
 
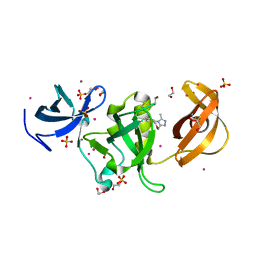 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | 分子名称: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | 著者 | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-08-30 | | 公開日 | 2017-10-11 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
6BPI
 
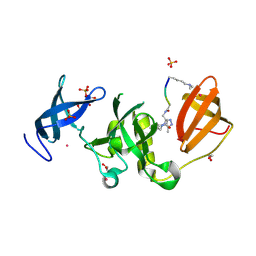 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | 分子名称: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | 著者 | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-11-23 | | 公開日 | 2017-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
3FLD
 
 | |
6N8H
 
 | |
6N8I
 
 | |
6N8F
 
 | |
6AU3
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | 分子名称: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, N-{[2-(3,5-dimethyl-4H-1,2,4-triazol-4-yl)phenyl]methyl}acetamide, ... | | 著者 | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-08-30 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
to be published
|
|
1XV0
 
 | | Solution NMR structure of RNA internal loop with three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCGAAGCCG | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P)-3', 5'-R(*GP*GP*UP*GP*GP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Znosko, B.M, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2004-10-26 | | 公開日 | 2004-11-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of an RNA Internal Loop with Three Consecutive Sheared GA Pairs
Biochemistry, 44, 2005
|
|
7RQ5
 
 | |
2DD1
 
 | | Three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCAAAGCCG | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*AP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*GP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2006-01-19 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
2DD3
 
 | | An alternating sheared AA pair in 5'GGUGAAGGCU/3'PCCGAAGCCG: II. The minor conformation with A6/A5/A16 stack | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*AP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2006-01-19 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
2DD2
 
 | | An alternating sheared AA pair in 5'GGUGAAGGCU/3'PCCGAAGCCG: I. The major conformation with A6/A15/A16 stack | | 分子名称: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*AP*AP*GP*GP*CP*U)-3' | | 著者 | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | 登録日 | 2006-01-19 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
1TUT
 
 | | J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns | | 分子名称: | 5'-R(*GP*AP*GP*GP*AP*AP*GP*GP*CP*GP*A)-3', 5'-R(*UP*CP*GP*UP*UP*AP*AP*UP*CP*UP*C)-3' | | 著者 | Znosko, B.M, Kennedy, S.D, Wille, P.C, Krugh, T.R, Turner, D.H. | | 登録日 | 2004-06-25 | | 公開日 | 2004-12-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Features and Thermodynamics of the J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns.
Biochemistry, 43, 2004
|
|
2KY0
 
 | |
2KY1
 
 | |
2KY2
 
 | |
2KXZ
 
 | |
2KQ6
 
 | | The structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity | | 分子名称: | Polycystin-2 | | 著者 | Petri, E.T, Celic, A, Kennedy, S.D, Ehrlich, B.E, Boggon, T.J, Hodsdon, M.E. | | 登録日 | 2009-10-28 | | 公開日 | 2010-05-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5V2R
 
 | |
2L8F
 
 | |
