3ZYN
 
 | | Crystal structure of the N-terminal leucine rich repeats of Netrin-G Ligand-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4B | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
3ZYJ
 
 | | NetrinG1 in complex with NGL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4C, ... | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
8Q7O
 
 | | Crystal structure of the FZD3 cysteine-rich domain in complex with a nanobody (14478) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J.S, Zhao, Y, Malinauskas, T, Jones, E.Y. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into Frizzled3 through nanobody modulators.
Nat Commun, 15, 2024
|
|
8QW4
 
 | | FZD3 in complex with nanobody 9 | | Descriptor: | Frizzled-3, Nanobody Nb9 | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into Frizzled3 through nanobody modulators.
Nat Commun, 15, 2024
|
|
4PBX
 
 | | Crystal structure of the six N-terminal domains of human receptor protein tyrosine phosphatase sigma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase S | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4PBW
 
 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4PBV
 
 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform, ... | | Authors: | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2014-04-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
3SUA
 
 | | Crystal structure of the intracellular domain of Plexin-B1 in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B1, ... | | Authors: | Bell, C.H, Aricescu, A.R, Jones, E.Y, Siebold, C. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.39 Å) | | Cite: | A Dual Binding Mode for RhoGTPases in Plexin Signalling.
Plos Biol., 9, 2011
|
|
3SU8
 
 | | Crystal structure of a truncated intracellular domain of Plexin-B1 in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B1, ... | | Authors: | Bell, C.H, Aricescu, A.R, Jones, E.Y, Siebold, C. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Dual Binding Mode for RhoGTPases in Plexin Signalling.
Plos Biol., 9, 2011
|
|
3X1O
 
 | | Crystal structure of the ROQ domain of human Roquin | | Descriptor: | IODIDE ION, Roquin-1 | | Authors: | Ose, T, Verma, A, Cockburn, J.B, Berrow, N.S, Alderton, D, Stuart, D, Owens, R.J, Jones, E.Y. | | Deposit date: | 2014-11-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis
Nat Commun, 6, 2015
|
|
5FWW
 
 | | Wnt modulator Kremen in complex with DKK1 (CRD2) and LRP6 (PE3PE4) | | Descriptor: | CALCIUM ION, DICKKOPF-RELATED PROTEIN 1, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWT
 
 | | Wnt modulator Kremen crystal form I at 2.10A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
6QP7
 
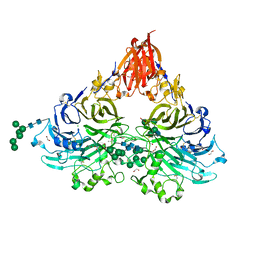 | | Drosophila Semaphorin 2a | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6QP8
 
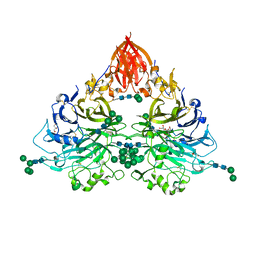 | | Drosophila Semaphorin 2b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6QP9
 
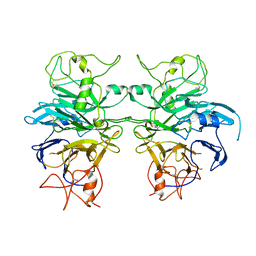 | | Drosophila Semaphorin 1a, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin-1A | | Authors: | Rozbesky, D, Robinson, R.A, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6XTZ
 
 | |
6YUW
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | Descriptor: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV2
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 598 | | Descriptor: | (3~{R})-1-phenylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YXI
 
 | | Structure of Notum in complex with a 1-(3-Chlorophenyl)-2,5-dimethyl-1H-pyrrole-3-carboxylic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-chlorophenyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchia, L, Jones, E.Y, Ruza, R.R, Hillier, J, Zhao, Y. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV4
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 686 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV0
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 587 | | Descriptor: | (3~{R})-1-(2-chlorophenyl)pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YUY
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 471 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-5-(trifluoromethyl)-1~{H}-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YSK
 
 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | Descriptor: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6R8Q
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A BENZOTRIAZOLE FRAGMENT | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Vecchia, L, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
6R8R
 
 | | Structure of the Wnt deacylase Notum in complex with isoquinoline 45 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Zhao, Y, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
