4BJ8
 
 | | Zebavidin | | Descriptor: | BIOTIN, GLYCEROL, ZEBAVIDIN | | Authors: | Airenne, T.T, Parthiban, M, Niederhauser, B, Zmurko, J, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S. | | Deposit date: | 2013-04-17 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zebavidin
Plos One, 8, 2013
|
|
4BCS
 
 | | Crystal structure of an avidin mutant | | Descriptor: | ACETATE ION, BIOTIN, CHIMERIC AVIDIN, ... | | Authors: | Airenne, T.T, Niederhauser, B, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel Chimeric Avidin with Increased Thermal Stability Using DNA Shuffling.
Plos One, 9, 2014
|
|
4BBO
 
 | | Crystal structure of core-bradavidin | | Descriptor: | ACETATE ION, BIOTIN, BLR5658 PROTEIN, ... | | Authors: | Airenne, T.T, Johnson, M.S, Maatta, J.A.E, Hytonen, V.H, Kulomaa, M.S. | | Deposit date: | 2012-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Core-Bradavidin
To be Published
|
|
5LUR
 
 | | An avidin mutant | | Descriptor: | Avidin, CHLORIDE ION, PROGESTERONE | | Authors: | Agrawal, N, Lehtonen, S.I, Riihimaki, T.A, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An avidin mutant
TO BE PUBLISHED
|
|
8TLM
 
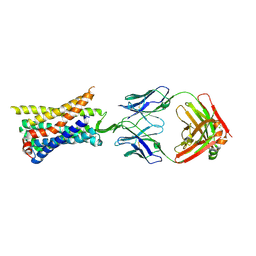 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8U1U
 
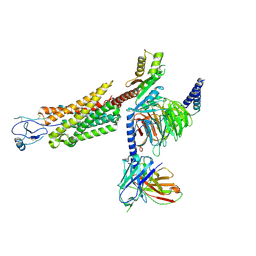 | | Structure of a class A GPCR/agonist complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
1Q8I
 
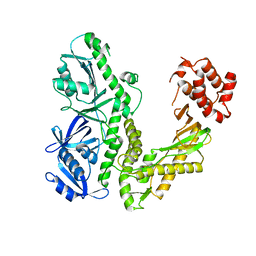 | | Crystal structure of ESCHERICHIA coli DNA Polymerase II | | Descriptor: | DNA polymerase II | | Authors: | Brunzelle, J.S, Muchmore, C.R.A, Mashhoon, N, Blair-Johnson, M, Shuvalova, L, Goodman, M.F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia Coli DNA Polymerase II
To be Published
|
|
1DNW
 
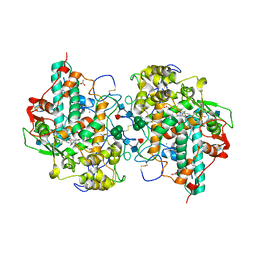 | | HUMAN MYELOPEROXIDASE-CYANIDE-THIOCYANATE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CALCIUM ION, ... | | Authors: | Blair-Johnson, M, Fiedler, T.J, Fenna, R.E. | | Deposit date: | 1999-12-16 | | Release date: | 2001-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human myeloperoxidase: structure of a cyanide complex and its interaction with bromide and thiocyanate substrates at 1.9 A resolution.
Biochemistry, 40, 2001
|
|
1DNU
 
 | | STRUCTURAL ANALYSES OF HUMAN MYELOPEROXIDASE-THIOCYANATE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CALCIUM ION, ... | | Authors: | Blair-Johnson, M, Fiedler, T.J, Fenna, R.E. | | Deposit date: | 1999-12-16 | | Release date: | 2001-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human myeloperoxidase: structure of a cyanide complex and its interaction with bromide and thiocyanate substrates at 1.9 A resolution.
Biochemistry, 40, 2001
|
|
1US1
 
 | | Crystal structure of human vascular adhesion protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Airenne, T.T, Nymalm, Y, Kidron, H, Soderholm, A, Johnson, M.S, Salminen, T.A. | | Deposit date: | 2003-11-17 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Human Vascular Adhesion Protein-1: Unique Structural Features with Functional Implications.
Protein Sci., 14, 2005
|
|
3RKZ
 
 | | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor capable of significantly decreasing tumor volume in a mouse xenograft model. | | Descriptor: | (5R,6S)-3-(L-alanyl)-5,6,15,15,18-pentamethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-12,8-(metheno)[1,5,9]triazacyclotetradecino[1,2-a]indole-9-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, Li, Z, Dushin, R.G, Nittoli, T, Otteng, M, Nikitenko, A, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E, Olland, A, Johnson, M, Levin, J.I. | | Deposit date: | 2011-04-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5693 Å) | | Cite: | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor which significantly decreases tumor volume in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2JXR
 
 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
7LBE
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBG
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBF
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
2C1S
 
 | | X-ray structure of biotin binding protein from chicken | | Descriptor: | BIOTIN BINDING PROTEIN A, BIOTIN-D-SULFOXIDE | | Authors: | Hytonen, V.P, Niskanen, E.A, Maatta, J.A.E, Huuskonen, J, Helttunen, K.J, Halling, K.K, Slotte, J.P, Nordlund, H.R, Rissanen, K, Johnson, M.S, Salminen, T.A, Kulomaa, M.S, Laitinen, O.H, Airenne, T.T. | | Deposit date: | 2005-09-19 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Characterization of a Novel Chicken Biotin-Binding Protein a (Bbp-A).
Bmc Struct.Biol., 7, 2007
|
|
2C1Q
 
 | | X-ray structure of biotin binding protein from chicken | | Descriptor: | BIOTIN, BIOTIN BINDING PROTEIN A, GLYCEROL | | Authors: | Hytonen, V.P, Niskanen, E.A, Maatta, J.A.E, Huuskonen, J, Helttunen, K.J, Halling, K.K, Slotte, J.P, Nordlund, H.R, Rissanen, K, Johnson, M.S, Salminen, T.A, Kulomaa, M.S, Laitinen, O.H, Airenne, T.T. | | Deposit date: | 2005-09-19 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Characterization of a Novel Chicken Biotin-Binding Protein a (Bbp-A).
Bmc Struct.Biol., 7, 2007
|
|
2Y32
 
 | | Crystal structure of bradavidin | | Descriptor: | BLR5658 PROTEIN | | Authors: | Leppiniemi, J, Gronroos, T, Johnson, M.S, Kulomaa, M.S, Hytonen, V.P, Airenne, T.T. | | Deposit date: | 2010-12-17 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Bradavidin - C-Terminal Residues Act as Intrinsic Ligands.
Plos One, 7, 2012
|
|
2C4I
 
 | | Crystal structure of engineered avidin | | Descriptor: | AVIDIN, BIOTIN, SULFATE ION | | Authors: | Hytonen, V.P, Horha, J, Airenne, T.T, Niskanen, E.A, Helttunen, K, Johnson, M.S, Salminen, T.A, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Controlling Quaternary Structure Assembly: Subunit Interface Engineering and Crystal Structure of Dual Chain Avidin.
J.Mol.Biol., 359, 2006
|
|
2UZ2
 
 | | Crystal structure of Xenavidin | | Descriptor: | ACETATE ION, BIOTIN, XENAVIDIN | | Authors: | Helppolainen, S.H, Maatta, J.A.E, Airenne, T.T, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-04-24 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characteristics of Xenavidin, the First Frog Avidin from Xenopus Tropicalis.
Bmc Struct.Biol., 9, 2009
|
|
1F6L
 
 | | VARIABLE LIGHT CHAIN DIMER OF ANTI-FERRITIN ANTIBODY | | Descriptor: | ANTI-FERRITIN IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Nymalm, Y, Kravchuk, Z, Salminen, T, Chumanevich, A.A, Dubnovitsky, A.P, Kankare, J, Pentikainen, O, Lehtonen, J, Arosio, P, Martsev, S, Johnson, M.S. | | Deposit date: | 2000-06-22 | | Release date: | 2002-10-23 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antiferritin VL homodimer binds human spleen ferritin with high specificity
J.STRUCT.BIOL., 138, 2002
|
|
2UYW
 
 | | Crystal structure of Xenavidin | | Descriptor: | BIOTIN, FORMIC ACID, XENAVIDIN | | Authors: | Helppolainen, S.H, Maatta, J.A.E, Airenne, T.T, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-04-20 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characteristics of Xenavidin, the First Frog Avidin from Xenopus Tropicalis.
Bmc Struct.Biol., 9, 2009
|
|
1PT6
 
 | | I domain from human integrin alpha1-beta1 | | Descriptor: | GLYCEROL, Integrin alpha-1, MAGNESIUM ION | | Authors: | Nymalm, Y, Puranen, J.S, Nyholm, T.K.M, Kapyla, J, Kidron, H, Pentikainen, O, Airenne, T.T, Heino, J, Slotte, J.P, Johnson, M.S, Salminen, T.A. | | Deposit date: | 2003-06-23 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Jararhagin-derived RKKH peptides induce structural changes in alpha1I domain of human integrin alpha1beta1.
J.Biol.Chem., 279, 2004
|
|
1QCY
 
 | | THE CRYSTAL STRUCTURE OF THE I-DOMAIN OF HUMAN INTEGRIN ALPHA1BETA1 | | Descriptor: | I-DOMAIN OF INTEGRIN ALPHA1BETA1, MAGNESIUM ION | | Authors: | Kankare, J.A, Salminen, T.A, Nymalm, Y, Kaepylae, J, Heino, J, Johnson, M.S. | | Deposit date: | 1999-05-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Jararhagin-derived RKKH Peptides Induce Structural Changes in a1I Domain of Human Integrin a1b1
J.Biol.Chem., 279, 2004
|
|
2ZM3
 
 | | Complex Structure of Insulin-like Growth Factor Receptor and Isoquinolinedione Inhibitor | | Descriptor: | (4Z)-6-bromo-4-({[4-(pyrrolidin-1-ylmethyl)phenyl]amino}methylidene)isoquinoline-1,3(2H,4H)-dione, Insulin-like growth factor 1 receptor | | Authors: | Xu, W, Mayer, S.C, Boschelli, F, Johnson, M, Dwyer, B. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead identification to generate isoquinolinedione inhibitors of insulin-like growth factor receptor (IGF-1R) for potential use in cancer treatment
Bioorg.Med.Chem.Lett., 18, 2008
|
|
