1E5A
 
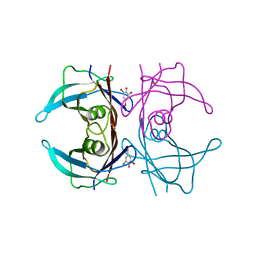 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | 2,4,6-TRIBROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3HY2
 
 | | Crystal Structure of Sulfiredoxin in Complex with Peroxiredoxin I and ATP:Mg2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxiredoxin-1, ... | | Authors: | Jonsson, T.J, Johnson, L.C, Lowther, W.T. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-06 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein Engineering of the Quaternary Sulfiredoxin-Peroxiredoxin Enzyme-Substrate Complex Reveals the Molecular Basis for Cysteine Sulfinic Acid Phosphorylation
J.Biol.Chem., 284, 2009
|
|
3CYI
 
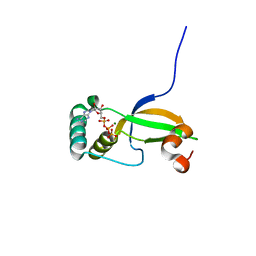 | | Crystal Structure of Human Sulfiredoxin (Srx) in Complex with ATP:Mg2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Sulfiredoxin-1 | | Authors: | Jonsson, T.J, Murray, M.S, Johnson, L.C, Lowther, W.T. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reduction of Cysteine Sulfinic Acid in Peroxiredoxin by Sulfiredoxin Proceeds Directly through a Sulfinic Phosphoryl Ester Intermediate.
J.Biol.Chem., 283, 2008
|
|
1QIC
 
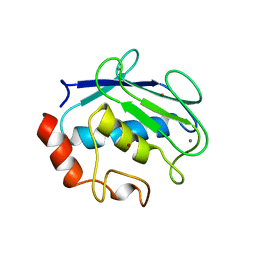 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
1QIA
 
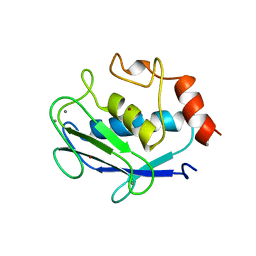 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
3G7A
 
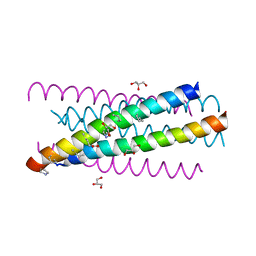 | | HIV gp41 six-helix bundle composed of a chimeric alpha+alpha/beta-peptide analogue of the CHR domain in complex with an NHR domain alpha-peptide | | Descriptor: | ACETYL GROUP, Chimeric alpha+alpha/beta-peptide analogue of the HIV gp41 CHR domain, Envelope glycoprotein gp160, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biological mimicry of protein surface recognition by alpha/beta-peptide foldamers
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1QIB
 
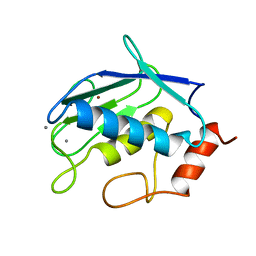 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
2GPB
 
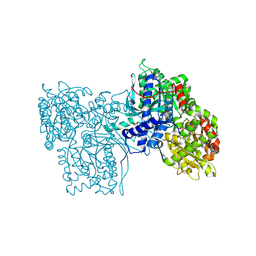 | |
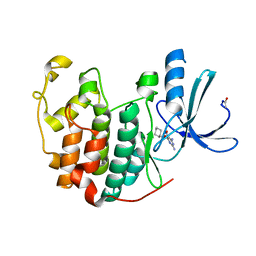
1E1X
 
 | |
1E3F
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-06-14 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E1V
 
 | |
1E4O
 
 | | Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phosphorylase Recognition and Phosphorylysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
1FQ1
 
 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1H0V
 
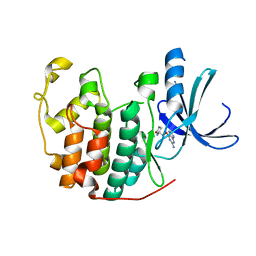 | | Human cyclin dependent protein kinase 2 in complex with the inhibitor 2-Amino-6-[(R)-pyrrolidino-5'-yl]methoxypurine | | Descriptor: | 5-{[(2-AMINO-9H-PURIN-6-YL)OXY]METHYL}-2-PYRROLIDINONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Gibson, A.E, Arris, C.E, Bentley, J, Boyle, F.T, Curtin, N.J, Davies, T.G, Endicott, J.A, Golding, B.T, Grant, S, Griffin, R.J, Jewsbury, P, Johnson, L.N, Mesguiche, V, Newell, D.R, Noble, M.E.M, Tucker, J.A, Whitfield, H.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
1HCJ
 
 | |
1H0W
 
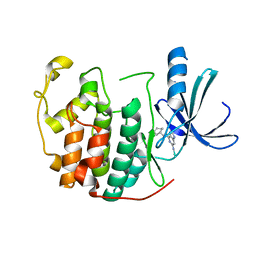 | | Human cyclin dependent protein kinase 2 in complex with the inhibitor 2-Amino-6-[cyclohex-3-enyl]methoxypurine | | Descriptor: | 1-AMINO-6-CYCLOHEX-3-ENYLMETHYLOXYPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Gibson, A.E, Arris, C.E, Bentley, J, Boyle, F.T, Curtin, N.J, Davies, T.G, Endicott, J.A, Golding, B.T, Grant, S, Griffin, R.J, Jewsbury, P, Johnson, L.N, Mesguiche, V, Newell, D.R, Noble, M.E.M, Tucker, J.A, Whitfield, H.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
1PHK
 
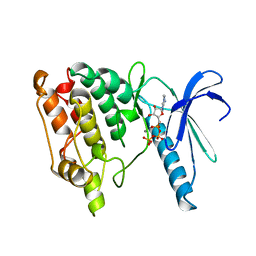 | | TWO STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHORYLASE, KINASE: AN ACTIVE PROTEIN KINASE COMPLEXED WITH NUCLEOTIDE, SUBSTRATE-ANALOGUE AND PRODUCT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE | | Authors: | Owen, D.J, Noble, M.E.M, Garman, E.F, Papageorgiou, A.C, Johnson, L.N. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
Structure, 3, 1995
|
|
1QL6
 
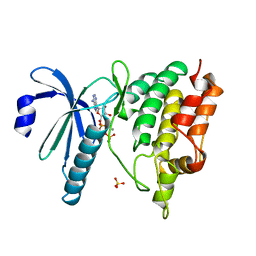 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
1QM5
 
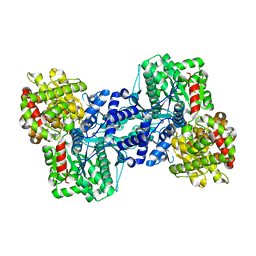 | | Phosphorylase recognition and phosphorylysis of its oligosaccharide substrate: answers to a long outstanding question | | Descriptor: | 4-thio-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | Deposit date: | 1999-09-20 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylase Recognition and Phosphorolysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
1TAH
 
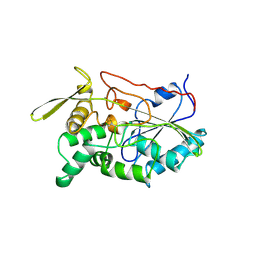 | | THE CRYSTAL STRUCTURE OF TRIACYLGLYCEROL LIPASE FROM PSEUDOMONAS GLUMAE REVEALS A PARTIALLY REDUNDANT CATALYTIC ASPARTATE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Noble, M.E.M, Cleasby, A, Johnson, L.N, Egmond, M, Frenken, L.G.J. | | Deposit date: | 1993-12-21 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate.
FEBS Lett., 331, 1993
|
|
2PRI
 
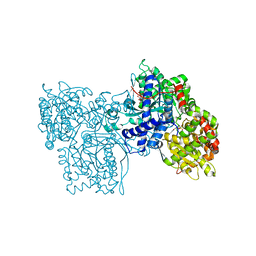 | | BINDING OF 2-DEOXY-GLUCOSE-6-PHOSPHATE TO GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLYCOGEN PHOSPHORYLASE B, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Zographos, S.E, Johnson, L.N, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The binding of 2-deoxy-D-glucose 6-phosphate to glycogen phosphorylase b: kinetic and crystallographic studies.
J.Mol.Biol., 254, 1995
|
|
1Q4K
 
 | | The polo-box domain of Plk1 in complex with a phospho-peptide | | Descriptor: | Phospho-peptide sequence Met.Gln.Ser.pThr.Pro.Leu, Serine/threonine-protein kinase PLK | | Authors: | Cheng, K, Lowe, E.D, Sinclair, J, Nigg, E.A, Johnson, L.N. | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex.
Embo J., 22, 2003
|
|
2PRJ
 
 | | Binding of N-acetyl-beta-D-glucopyranosylamine to Glycogen Phosphorylase B | | Descriptor: | GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kontou, M, Zographos, S.E, Watson, K.A, Johnson, L.N, Bichard, C.J.F, Fleet, G.W.J, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-acetyl-beta-D-glucopyranosylamine: a potent T-state inhibitor of glycogen phosphorylase. A comparison with alpha-D-glucose.
Protein Sci., 4, 1995
|
|
2PHK
 
 | | THE CRYSTAL STRUCTURE OF A PHOSPHORYLASE KINASE PEPTIDE SUBSTRATE COMPLEX: KINASE SUBSTRATE RECOGNITION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lowe, E.D, Noble, M.E.M, Skamnaki, V.T, Oikonomakos, N.G, Owen, D.J, Johnson, L.N. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition.
EMBO J., 16, 1997
|
|
1QMZ
 
 | | PHOSPHORYLATED CDK2-CYCLYIN A-SUBSTRATE PEPTIDE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN A, ... | | Authors: | Brown, N.R, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 1999-10-11 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for Specificity of Substrate and Recruitment Peptides for Cyclin-Dependent Kinases
Nat.Cell Biol., 1, 1999
|
|
