1F8V
 
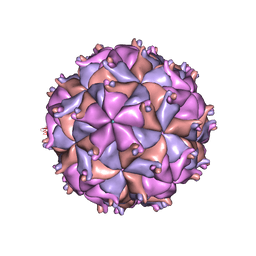 | | THE STRUCTURE OF PARIACOTO VIRUS REVEALS A DODECAHEDRAL CAGE OF DUPLEX RNA | | Descriptor: | CALCIUM ION, MATURE CAPSID PROTEIN BETA, MATURE CAPSID PROTEIN GAMMA, ... | | Authors: | Tang, L, Johnson, K.N, Ball, L.A, Lin, T, Yeager, M, Johnson, J.E. | | Deposit date: | 2000-07-05 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA.
Nat.Struct.Biol., 8, 2001
|
|
7RXS
 
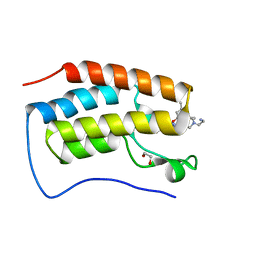 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
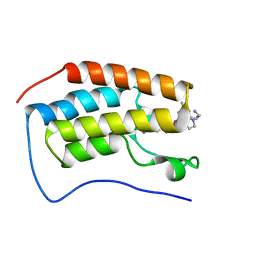 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXR
 
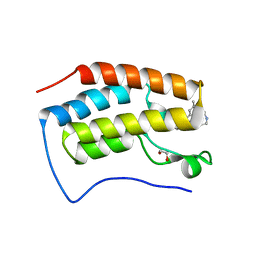 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
3J31
 
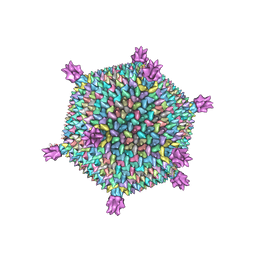 | | Life in the extremes: atomic structure of Sulfolobus Turreted Icosahedral Virus | | Descriptor: | A223 penton base, A55 membrane protein, C381 turret protein, ... | | Authors: | Veesler, D, Ng, T.S, Sendamarai, A.K, Eilers, B.J, Lawrence, C.M, Lok, S.M, Young, M.J, Johnson, J.E, Fu, C.-Y. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3IYI
 
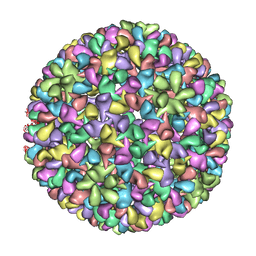 | | P22 expanded head coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | P22 coat protein in procapsid shells | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
8A41
 
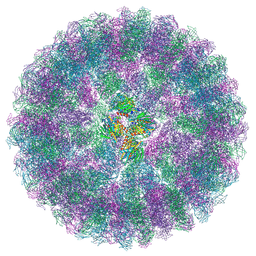 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A3C
 
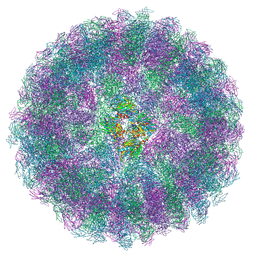 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A6J
 
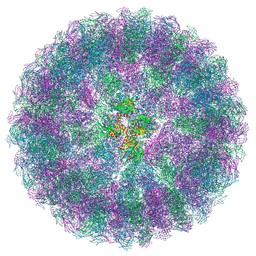 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
3IYH
 
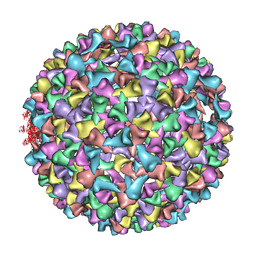 | | P22 procapsid coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | Coat protein | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
8AC6
 
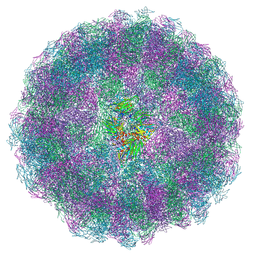 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): medium class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8ACH
 
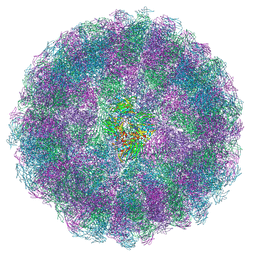 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8AAY
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): small class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
1NY7
 
 | | COWPEA MOSAIC VIRUS (CPMV) | | Descriptor: | COWPEA MOSAIC VIRUS, LARGE (L) SUBUNIT, SMALL (S) SUBUNIT | | Authors: | Lin, T, Chen, Z, Usha, R, Stauffacher, C.V, Dai, J.-B, Schmidt, T, Johnson, J.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Refined Crystal Structure of Cowpea Mosaic Virus at 2.8A Resolution
Virology, 265, 1999
|
|
1PGL
 
 | | BEAN POD MOTTLE VIRUS (BPMV), MIDDLE COMPONENT | | Descriptor: | 5'-R(*AP*GP*UP*CP*UP*C)-3', BEAN POD MOTTLE VIRUS LARGE (L) SUBUNIT, BEAN POD MOTTLE VIRUS SMALL (S) SUBUNIT | | Authors: | Lin, T, Cavarelli, J, Johnson, J.E. | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for assembly-dependent folding of protein and RNA in an icosahedral virus.
Virology, 314, 2003
|
|
1PGW
 
 | | BEAN POD MOTTLE VIRUS (BPMV), TOP COMPONENT | | Descriptor: | BEAN POD MOTTLE VIRUS LARGE (L) SUBUNIT, BEAN POD MOTTLE VIRUS SMALL (S) SUBUNIT | | Authors: | Lin, T, Cavarelli, J, Johnson, J.E. | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence for assembly-dependent folding of protein and RNA in an icosahedral virus.
Virology, 314, 2003
|
|
1PEH
 
 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPNH1 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
1PEI
 
 | | NMR STRUCTURE OF THE MEMBRANE-BINDING DOMAIN OF CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE, 10 STRUCTURES | | Descriptor: | PEPC22 | | Authors: | Dunne, S.J, Cornell, R.B, Johnson, J.E, Glover, N.R, Tracey, A.S. | | Deposit date: | 1996-06-10 | | Release date: | 1996-12-07 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase.
Biochemistry, 35, 1996
|
|
3BM9
 
 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | Descriptor: | 4-bromo-6-(6-hydroxy-1,2-benzisoxazol-3-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | Deposit date: | 2007-12-12 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
1IF0
 
 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
7RSK
 
 | | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein | | Authors: | Kim, Y, Nocek, B, Endres, M, Joachimiak, G, Johnson, J, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus
To Be Published
|
|
7KW7
 
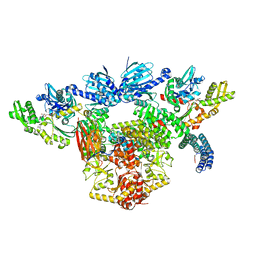 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | Authors: | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
3BMY
 
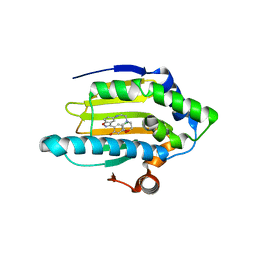 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | Descriptor: | 4-chloro-6-{5-[(2-morpholin-4-ylethyl)amino]-1,2-benzisoxazol-3-yl}benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
1RNA
 
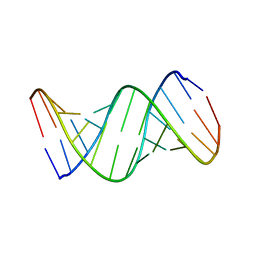 | | CRYSTALLOGRAPHIC STRUCTURE OF AN RNA HELIX: [U(U-A)6A]2 | | Descriptor: | RNA (5'-R(*UP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*A)-3') | | Authors: | Dock-Bregeon, A.C, Chevrier, B, Podjarny, A, Johnson, J, De Bear, J.S, Gough, G.R, Gilham, P.T, Moras, D. | | Deposit date: | 1990-02-01 | | Release date: | 1991-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic structure of an RNA helix: [U(UA)6A]2.
J.Mol.Biol., 209, 1989
|
|
7K3M
 
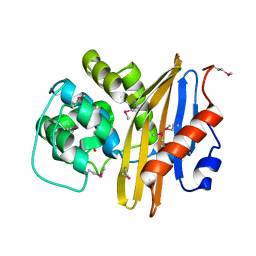 | | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Sherrell, D.A, Johnson, J, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography
To Be Published
|
|
