5E3E
 
 | | Crystal structure of CdiA-CT/CdiI complex from Y. kristensenii 33638 | | Descriptor: | CdiI immunity protein, Large exoprotein involved in heme utilization or adhesion, SODIUM ION | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily.
Nucleic Acids Res., 45, 2017
|
|
6NBG
 
 | | 2.05 Angstrom Resolution Crystal Structure of Hypothetical Protein KP1_5497 from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Glucosamine-6-phosphate deaminase, PHOSPHATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6WTC
 
 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
6WHL
 
 | |
6CX8
 
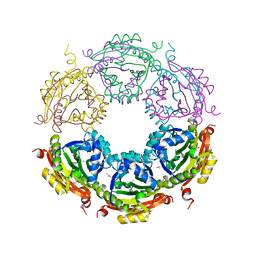 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, MANGANESE (II) ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae in complex with manganese ions.
To Be Published
|
|
1NG5
 
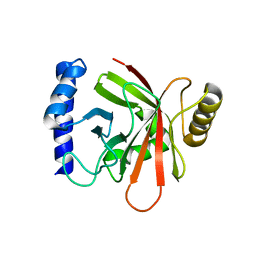 | |
4DGH
 
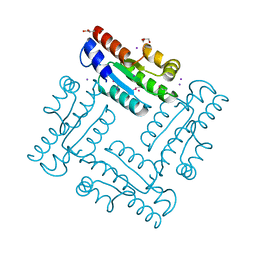 | | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution | | Descriptor: | GLYCEROL, IODIDE ION, POTASSIUM ION, ... | | Authors: | Keller, J.P, Chang, C, Marshall, N, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution
To be Published
|
|
1P99
 
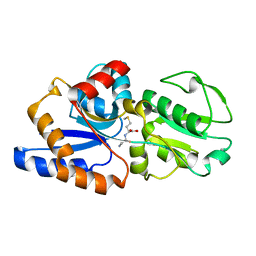 | | 1.7A crystal structure of protein PG110 from Staphylococcus aureus | | Descriptor: | GLYCINE, Hypothetical protein PG110, METHIONINE | | Authors: | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions.
Biochemistry, 43, 2004
|
|
2R41
 
 | |
2QRR
 
 | | Crystal structure of the soluble domain of the ABC transporter, ATP-binding protein from Vibrio parahaemolyticus | | Descriptor: | CHLORIDE ION, Methionine import ATP-binding protein metN | | Authors: | Kim, Y, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-28 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Soluble Domain of the ABC Transporter, ATP-binding Protein from Vibrio parahaemolyticus.
To be Published
|
|
3GE2
 
 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
2RE1
 
 | | Crystal structure of aspartokinase alpha and beta subunits | | Descriptor: | Aspartokinase, alpha and beta subunits, CALCIUM ION | | Authors: | Chang, C, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of aspartokinase alpha and beta subunits.
To be Published
|
|
2R5R
 
 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
4DU6
 
 | | Crystal structure of GTP cyclohydrolase I from Yersinia pestis complexed with GTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Crystal structure of GTP cyclohydrolase I from Yersinia pestis complexed with GTP
To be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | Authors: | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-09-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
3BJB
 
 | | Crystal structure of a TetR transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | Probable transcriptional regulator, TetR family protein, SULFATE ION | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-03 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a TetR transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3TSN
 
 | | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, UNKNOWN LIGAND | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni.
To be Published
|
|
6U10
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, L-CAPTOPRIL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril.
To Be Published
|
|
6UAG
 
 | |
3BQY
 
 | | Crystal structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2). | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Putative TetR family transcriptional regulator | | Authors: | Cuff, M.E, Skarina, T, Kagan, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
TO BE PUBLISHED
|
|
6U2Z
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, 1,2-ETHANEDIOL, COPPER (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions
To Be Published
|
|
3C07
 
 | | Crystal structure of a TetR family transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | Putative tetR-family transcriptional regulator, SULFATE ION | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
To be Published
|
|
6V70
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Cadmium in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CADMIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
6MGZ
 
 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NDM-4, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae
To Be Published
|
|
6B8W
 
 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
