7QR1
 
 | | SpCas9 bound to TRAC off-target2 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR7
 
 | | SpCas9 bound to AAVS1 off-target2 DNA substrate | | Descriptor: | AAVS1 off-target2 non-target strand, AAVS1 off-target2 target strand, AAVS1 sgRNA, ... | | Authors: | Pacesa, M, JInek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
6F9N
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CPSF160-WDR33 COMPLEX | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, pre-mRNA 3' end processing protein WDR33 | | Authors: | Clerici, M, Jinek, M. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex.
Elife, 6, 2017
|
|
6FUW
 
 | | Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the PAS AAUAAA motif at 3.1 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*GP*G)-3'), ... | | Authors: | Clerici, M, Faini, M, Jinek, M. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7OX8
 
 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
8BD5
 
 | | Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, DNA non-target strand, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8BD4
 
 | | TniQ-capped Tns-ATP-dsDNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Querques, I, Schmitz, M, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8BD6
 
 | | Cas12k-sgRNA-dsDNA-TnsC non-productive complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cas12k, DNA, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
6O16
 
 | | Crystal structure of murine DHX37 in complex with RNA | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 37, RNA (5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Boneberg, F, Brandmann, T, Kobel, L, van den Heuvel, J, Bargsten, K, Bammert, L, Kutay, U, Jinek, M. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis.
Rna, 25, 2019
|
|
6F9W
 
 | |
6QZK
 
 | | Structure of Clostridium butyricum Argonaute bound to a guide DNA (5' deoxycytidine) and a 19-mer target DNA | | Descriptor: | Clostridium butyricum Argonaute, DNA target (5'-D(T*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*T)-3'), FORMIC ACID, ... | | Authors: | Swarts, D.C, Jinek, M, Hegge, J.W, Van der Oost, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.548 Å) | | Cite: | DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute.
Nucleic Acids Res., 47, 2019
|
|
8Q40
 
 | | Crystal structure of cA4 activated Can2 in complex with a cleaved DNA substrate | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*CP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q42
 
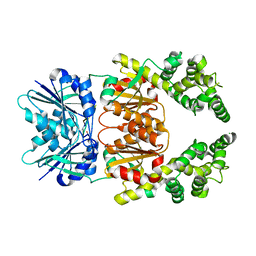 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-A DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*AP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q43
 
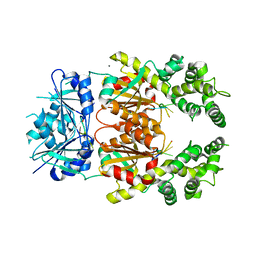 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-C DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*CP*CP*CP*CP*C)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q41
 
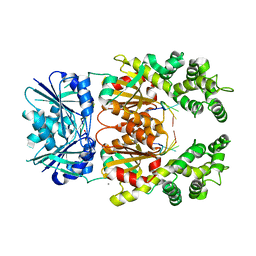 | | Crystal structure of Can2 (E341A) bound to cA4 and TTTAAA ssDNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q44
 
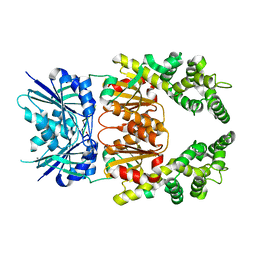 | | Crystal structure of cA4-bound Can2 (E364R) in complex with oligo-T DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*T)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q3Z
 
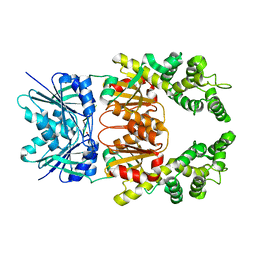 | |
8Q3Y
 
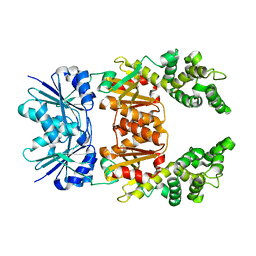 | |
6RW0
 
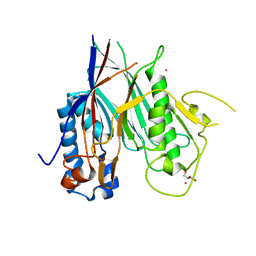 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase | | Descriptor: | GLYCEROL, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
6I0V
 
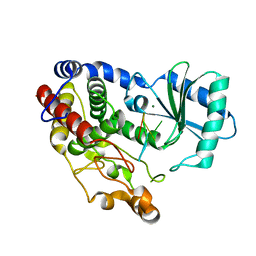 | | Crystal structure of DmTailor in complex with CACAGU RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*AP*CP*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
5NFV
 
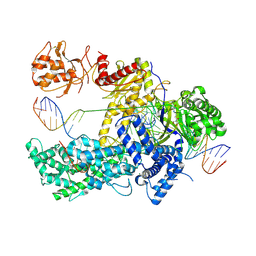 | | Crystal structure of catalytically inactive FnCas12 mutant bound to an R-loop structure containing a pre-crRNA mimic and full-length DNA target | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CRISPR-associated endonuclease Cpf1, DNA non-target strand, ... | | Authors: | Swarts, D.C, van der Oost, J, Jinek, M. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a.
Mol. Cell, 66, 2017
|
|
5FQ5
 
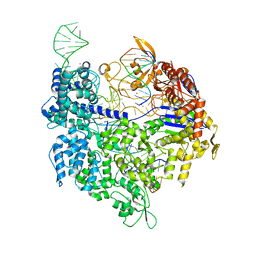 | | Crystal structure of Cas9-sgRNA-DNA complex solved by native SAD phasing | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Olieric, V, Weinert, T, Finke, A, Anders, C, Jinek, M, Wang, M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Data-Collection Strategy for Challenging Native Sad Phasing.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4BK0
 
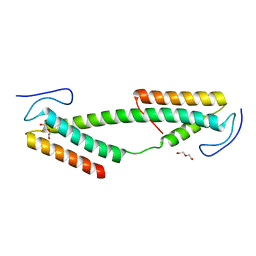 | | Crystal structure of the KIX domain of human RECQL5 (domain-swapped dimer) | | Descriptor: | ATP-DEPENDENT DNA HELICASE Q5, DI(HYDROXYETHYL)ETHER | | Authors: | Kassube, S.A, Jinek, M, Fang, J, Tsutakawa, S, Nogales, E. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mimicry in Transcription Regulation of Human RNA Polymerase II by the DNA Helicase Recql5
Nat.Struct.Mol.Biol., 20, 2013
|
|
