8X99
 
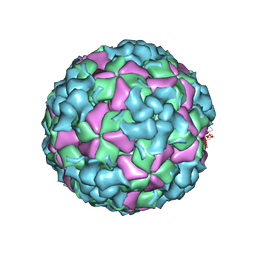 | | Cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab h1A6.2
To Be Published
|
|
8X95
 
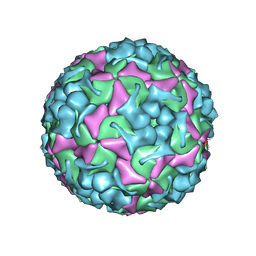 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2
To Be Published
|
|
8X97
 
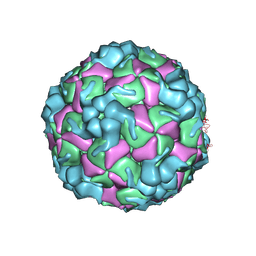 | | Cryo-EM structure of enterovirus A71 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structure of enterovirus A71 empty particle in complex with Fab h1A6.2
To Be Published
|
|
8X9A
 
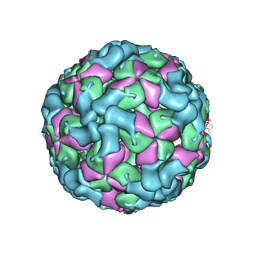 | | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2
To Be Published
|
|
7WDE
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, styrene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7WDI
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7WDD
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine, styrene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
5X74
 
 | | The crystal Structure PDE delta in complex with compound (R, R)-1g | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-[2-[4-[(2R)-2-(2-fluorophenyl)-4-oxidanylidene-1,2-dihydroquinazolin-3-yl]piperidin-1-yl]ethyl]-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5H1Y
 
 | |
8IBV
 
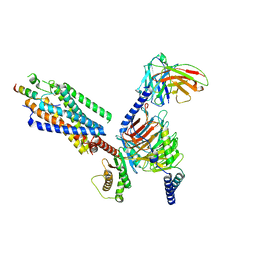 | | Cryo-EM structure of the motilin-bound motilin receptor-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Jiang, Y, Xu, H.E, You, C, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
4PQW
 
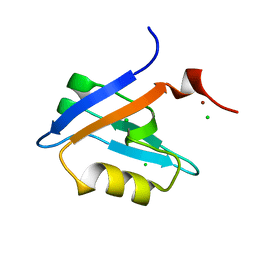 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
8WXB
 
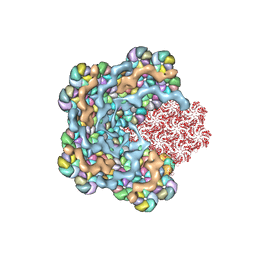 | | Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4 | | Descriptor: | Carboxysome assembly protein CsoS2, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1 | | Authors: | Jiang, Y.L, Zhou, R.Q, Zhou, C.Z, Zeng, Q.L. | | Deposit date: | 2023-10-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and assembly of the alpha-carboxysome in the marine cyanobacterium Prochlorococcus.
Nat.Plants, 10, 2024
|
|
3QFN
 
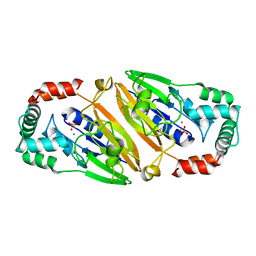 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH in complex with inorganic phosphate | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
3QFO
 
 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH im complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
2L5F
 
 | | Solution structure of the tandem WW domains from HYPA/FBP11 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interaction with polyglutamine expanded huntingtin alters cellular distribution and RNA processing of huntingtin yeast two-hybrid protein A (HYPA)
To be Published
|
|
3QFM
 
 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Putative uncharacterized protein | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
8HOT
 
 | |
8HOO
 
 | |
4MPA
 
 | | Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1, ... | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
7WDH
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDG
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4I5V
 
 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with Ap4A | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, BIS(ADENOSINE)-5'-TETRAPHOSPHATE | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
4I5W
 
 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with AMP | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, ADENOSINE MONOPHOSPHATE, PHOSPHATE ION | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
7EGN
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
