8SZE
 
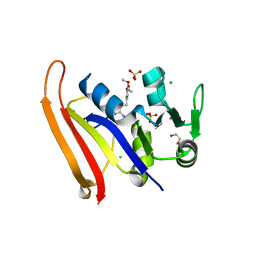 | | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim
To be published
|
|
1L4Y
 
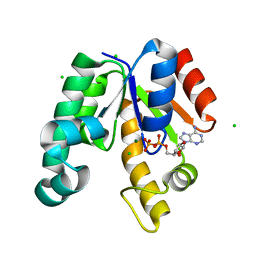 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AT 2.0 ANGSTROM RESOLUTION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
5Z0V
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
5Z0R
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
3FTD
 
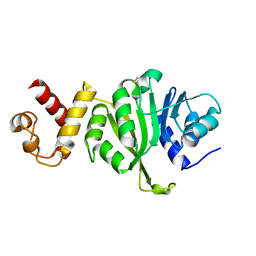 | |
3FTF
 
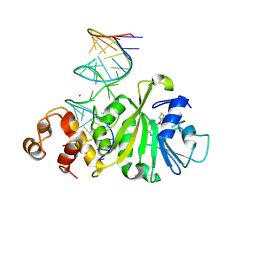 | | Crystal structure of A. aeolicus KsgA in complex with RNA and SAH | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase, POTASSIUM ION, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
3FTC
 
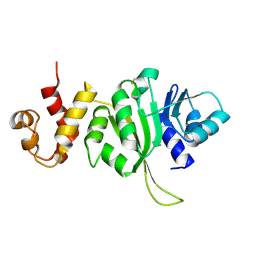 | |
3FTE
 
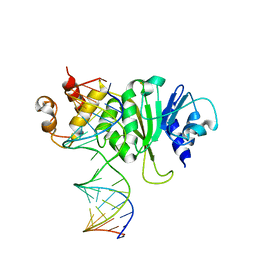 | | Crystal structure of A. aeolicus KsgA in complex with RNA | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
2LXH
 
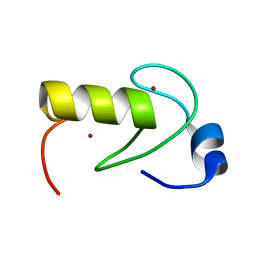 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2LXP
 
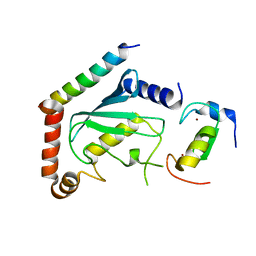 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
6PBZ
 
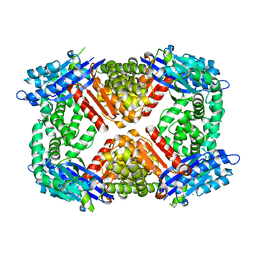 | | Crystal structure of Escherichia coli GppA | | Descriptor: | CHLORIDE ION, Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase | | Authors: | Song, H, Shaw, G.X, Wang, C, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
8H01
 
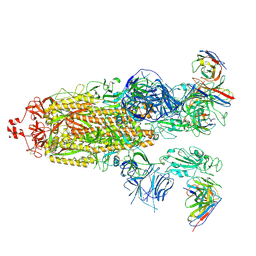 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8H00
 
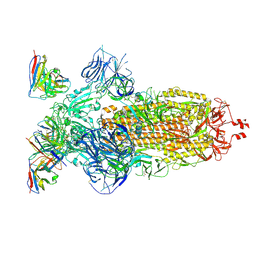 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
6PC3
 
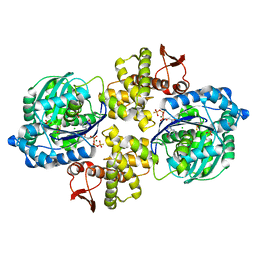 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GSP | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC2
 
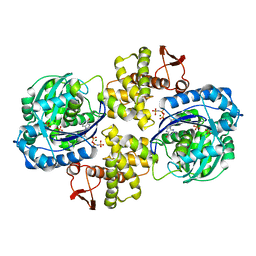 | | Crystal structure of Helicobacter pylori PPX/GppA in complex with GNP | | Descriptor: | Guanosine pentaphosphate phosphohydrolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
8ITU
 
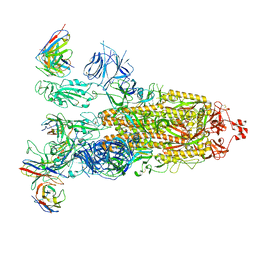 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 IgG. | | Descriptor: | 1H1 heavy chain, 1H1 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2023-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
5GJB
 
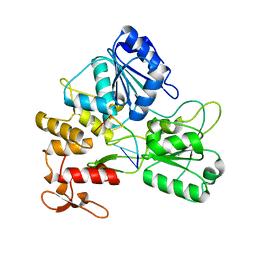 | | Zika virus NS3 helicase in complex with ssRNA | | Descriptor: | NS3 helicase, RNA (5'-R(*AP*GP*AP*UP*CP*AP*A)-3') | | Authors: | Tian, H.L, Ji, X.Y, Yang, X.Y, Zhang, Z.X, Lu, Z.K, Yang, K.L, Chen, C, Zhao, Q, Chi, H, Mu, Z.Y, Xie, W, Wang, Z.F, Lou, H.Q, Yang, H.T, Rao, Z.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis of Zika virus helicase in recognizing its substrates
Protein Cell, 7, 2016
|
|
5GJC
 
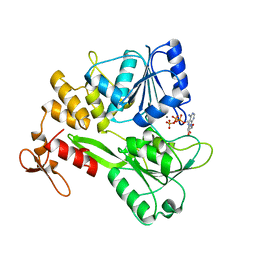 | | Zika virus NS3 helicase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, NS3 helicase | | Authors: | Tian, H.L, Ji, X.Y, Yang, X.Y, Zhang, Z.X, Lu, Z.K, Yang, K.L, Chen, C, Zhao, Q, Chi, H, Mu, Z.Y, Xie, W, Wang, Z.F, Lou, H.Q, Yang, H.T, Rao, Z.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of Zika virus helicase in recognizing its substrates
Protein Cell, 7, 2016
|
|
1RHH
 
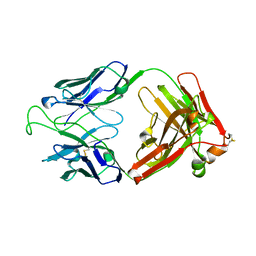 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
1TDI
 
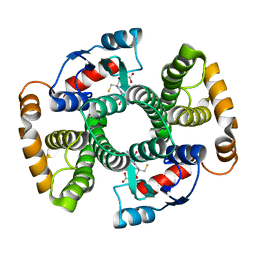 | | Crystal Structure of hGSTA3-3 in Complex with Glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A3-3 | | Authors: | Gu, Y, Guo, J, Pal, A, Pan, S.S, Zimniak, P, Singh, S.V, Ji, X. | | Deposit date: | 2004-05-22 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human glutathione S-transferase A3-3 and mechanistic implications for its high steroid isomerase activity.
Biochemistry, 43, 2004
|
|
1I4S
 
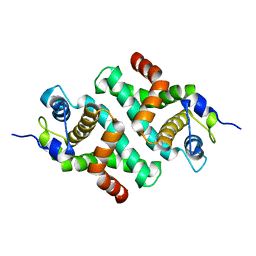 | |
3HD2
 
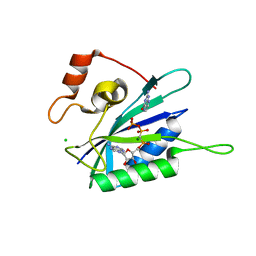 | | Crystal structure of E. coli HPPK(Q50A) in complex with MgAMPCPP and pterin | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
3HD1
 
 | | Crystal structure of E. coli HPPK(N10A) in complex with MgAMPCPP | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
3HSZ
 
 | | Crystal structure of E. coli HPPK(F123A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
3HT0
 
 | | Crystal structure of E. coli HPPK(F123A) in complex with MgAMPCPP | | Descriptor: | CHLORIDE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, HPPK, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
