3ZG8
 
 | | Crystal Structure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZPJ
 
 | | Crystal structure of Ton1535 from Thermococcus onnurineus NA1 | | Descriptor: | TON_1535 | | Authors: | Jeong, J.H, kim, Y.G. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structure of the Hypothetical Protein Ton1535 from Thermococcus Onnurineus Na1 Reveals Unique Structural Properties by a Left-Handed Helical Turn in Normal Alpha-Solenoid Protein.
Proteins, 82, 2014
|
|
4UOY
 
 | | Crystal structure of YgjG in complex with Pyridoxal-5'-phosphate | | Descriptor: | FORMIC ACID, GLYCEROL, PUTRESCINE AMINOTRANSFERASE, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2014-06-11 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Structure of Putrescine Aminotransferase from Escherichia Coli Provides Insights Into the Substrate Specificity Among Class III Aminotransferases.
Plos One, 9, 2014
|
|
4UOX
 
 | |
4UY5
 
 | |
4UY7
 
 | |
4UY6
 
 | |
2XMO
 
 | | The crystal structure of Lmo2642 | | Descriptor: | CALCIUM ION, FE (III) ION, LMO2642 PROTEIN, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2010-07-28 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analysis of the Lmo2642 Cyclic Nucleotide Phosphodiesterase from Listeria Monocytogenes.
Proteins, 79, 2011
|
|
5ZQB
 
 | |
5ZQA
 
 | |
5ZQC
 
 | | Crystal Structure of Penicillin-Binding Protein D2 from Listeria monocytogenes in the Ampicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Lmo2812 protein | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2018-04-18 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal Structures of Penicillin-Binding Protein D2 from Listeria monocytogenes and Structural Basis for Antibiotic Specificity
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZQE
 
 | | Crystal Structure of Penicillin-Binding Protein D2 from Listeria monocytogenes in the Cefuroxime bound form | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2018-04-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal Structures of Penicillin-Binding Protein D2 from Listeria monocytogenes and Structural Basis for Antibiotic Specificity
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZQD
 
 | |
4V3I
 
 | | Crystal Structure of TssL from Vibrio cholerae. | | Descriptor: | GLYCEROL, VCA0115 | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2014-10-19 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of the bacterial type VI secretion system component TssL from Vibrio cholerae.
J. Microbiol., 53, 2015
|
|
4ARZ
 
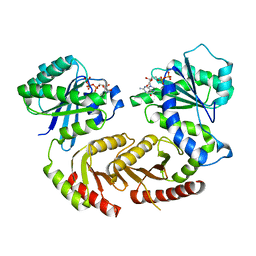 | | The crystal structure of Gtr1p-Gtr2p complexed with GTP-GDP | | Descriptor: | GTP-BINDING PROTEIN GTR1, GTP-BINDING PROTEIN GTR2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-04-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Gtr1Pgtp-Gtr2Pgdp Complex Reveals Large Structural Rearrangements Triggered by GTP-to-Gdp Conversion
J.Biol.Chem., 287, 2012
|
|
3ZG9
 
 | |
3ZGA
 
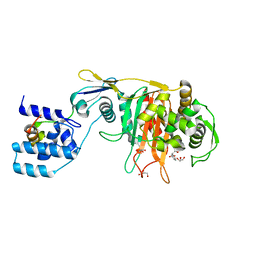 | | Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN 4 | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZG7
 
 | |
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | Descriptor: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | Authors: | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2014-09-04 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
4L7M
 
 | |
5XPC
 
 | | Crystal Structure of Drep4 CIDE domain | | Descriptor: | DNAation factor-related protein 4, GLYCEROL | | Authors: | Park, H.H, Jeong, J.H. | | Deposit date: | 2017-06-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | CIDE domains form functionally important higher-order assemblies for DNA fragmentation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4D2K
 
 | |
8I7Q
 
 | |
5G2G
 
 | |
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
