3BOE
 
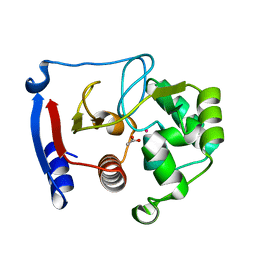 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 with acetate (CDCA1-R2) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
6PRK
 
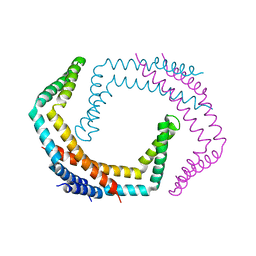 | | X-ray Crystal Structure of Bacillus subtilis RicA in complex with RicF | | Descriptor: | RicA, RicF | | Authors: | Khaja, F.T, Jeffrey, P.D, Neiditch, M.B, Dubnau, D. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Function Studies of the Bacillus subtilis Ric Proteins Identify the Fe-S Cluster-Ligating Residues and Their Roles in Development and RNA Processing.
Mbio, 10, 2019
|
|
8UBS
 
 | |
5VAE
 
 | |
5BV0
 
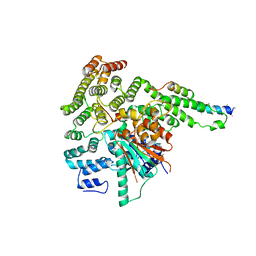 | |
5BUZ
 
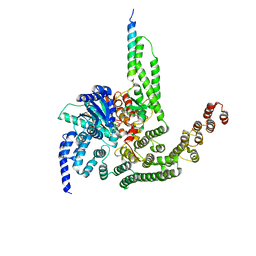 | |
5BV1
 
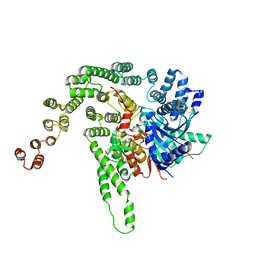 | |
5VAF
 
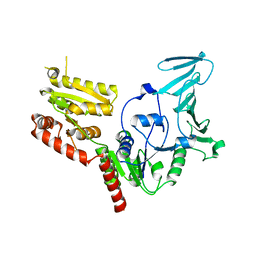 | |
8DIT
 
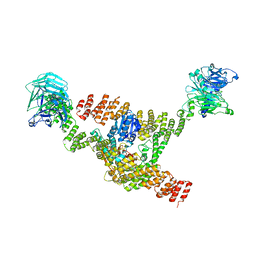 | | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | | Descriptor: | Vacuolar protein sorting-associated protein 16, Vacuolar protein sorting-associated protein 18, Vacuolar protein sorting-associated protein 33 | | Authors: | Port, S.A, Farrell, P.D, Jeffrey, P.D, DiMaio, F, Hughson, F.M. | | Deposit date: | 2022-06-29 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly
To Be Published
|
|
4CEV
 
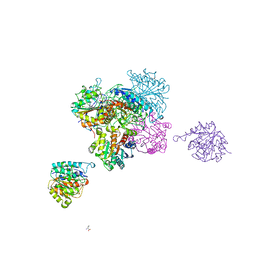 | | ARGINASE FROM BACILLUS CALDEVELOX, L-ORNITHINE COMPLEX | | Descriptor: | GUANIDINE, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
8CUK
 
 | |
1YY8
 
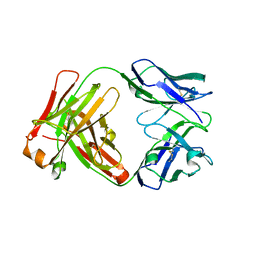 | | Crystal structure of the Fab fragment from the monoclonal antibody cetuximab/Erbitux/IMC-C225 | | Descriptor: | Cetuximab Fab Heavy chain, Cetuximab Fab Light chain | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
1YY9
 
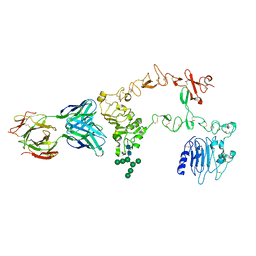 | | Structure of the extracellular domain of the epidermal growth factor receptor in complex with the Fab fragment of cetuximab/Erbitux/IMC-C225 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab Heavy chain, ... | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
6CFI
 
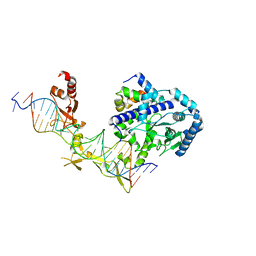 | | Crystal structure of Rad4-Rad23 bound to a 6-4 photoproduct UV lesion | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*(T64)P*TP*GP*GP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), DNA repair protein RAD4, DNA('-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*AP*AP*AP*GP*CP*TP*AP*CP*AP*A)-'), ... | | Authors: | Min, J, Jeffrey, P.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.36241913 Å) | | Cite: | Structure and mechanism of pyrimidine-pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex.
Nucleic Acids Res., 47, 2019
|
|
2VNM
 
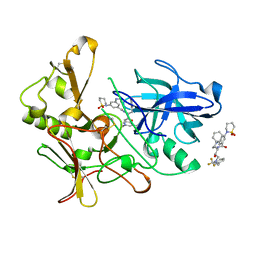 | | Human BACE-1 in complex with 3-(1,1-dioxidotetrahydro-2H-1,2-thiazin- 2-yl)-5-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-3-(1,1-dioxido-1,2-thiazinan-2-yl)-5-(ethylamino)benzamide | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, East, P, Hawkins, J, Howes, C, Hussain, I, Jeffrey, P, Maile, G, Matico, R, Mosley, J, Naylor, A, OBrien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2008-02-05 | | Release date: | 2008-05-20 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Second Generation of Hydroxyethylamine Bace-1 Inhibitors: Optimizing Potency and Oral Bioavailability.
J.Med.Chem., 51, 2008
|
|
2VNN
 
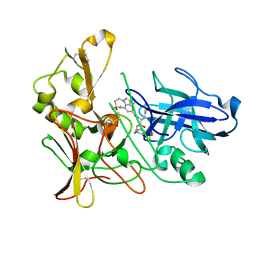 | | Human BACE-1 in complex with 7-ethyl-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)-1- methyl-3,4-dihydro-1H-(1,2,5)thiadiazepino(3,4,5-hi)indole-9- carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-7-ethyl-1-methyl-3,4-dihydro-1H-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide 2,2-dioxide | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, East, P, Hawkins, J, Howes, C, Hussain, I, Jeffrey, P, Maile, G, Matico, R, Mosley, J, Naylor, A, OBrien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2008-02-05 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Second Generation of Hydroxyethylamine Bace-1 Inhibitors: Optimizing Potency and Oral Bioavailability.
J.Med.Chem., 51, 2008
|
|
6D0H
 
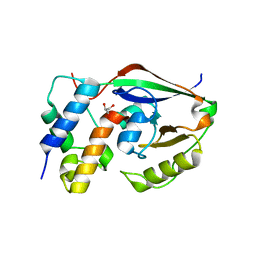 | |
6D0I
 
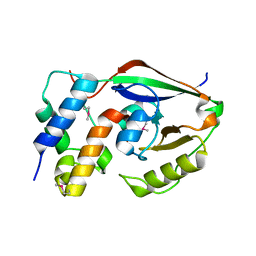 | | ParT: Prs ADP-ribosylating toxin bound to cognate antitoxin ParS. L48M ParT, SeMet-substituted complex. | | Descriptor: | GLYCEROL, ParS: COG5642 (DUF2384) antitoxin fragment, ParT: COG5654 (RES domain) toxin | | Authors: | Piscotta, F.J, Jeffrey, P.D, Link, A.J. | | Deposit date: | 2018-04-10 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ParST is a widespread toxin-antitoxin module that targets nucleotide metabolism.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6UFF
 
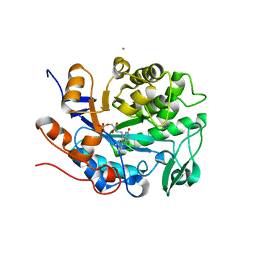 | |
6WC3
 
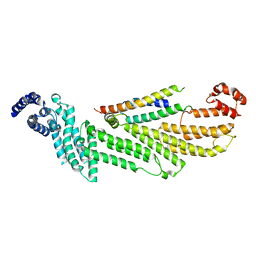 | |
8FTU
 
 | | Crystal structure of the SNARE Use1 bound to Dsl1 complex subunits Sec39 and Dsl1, Revised Use1 structure | | Descriptor: | Protein transport protein DSL1, Protein transport protein SEC39, Vesicle transport protein USE1 | | Authors: | Travis, S.M, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (5.73 Å) | | Cite: | Structure of a membrane tethering complex incorporating multiple SNAREs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1TSR
 
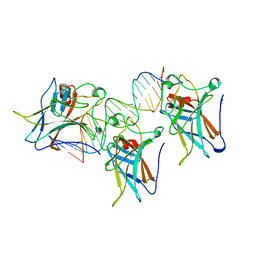 | | P53 CORE DOMAIN IN COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*TP*TP*GP*GP*GP*CP*AP*AP*GP*TP*CP*TP*A P*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*CP*CP*TP*AP*GP*AP*CP*TP*TP*GP*CP*CP*CP*A P*AP*TP*TP*A)-3'), PROTEIN (P53 TUMOR SUPPRESSOR), ... | | Authors: | Cho, Y, Gorina, S, Jeffrey, P, Pavletich, N. | | Deposit date: | 1995-07-28 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations.
Science, 265, 1994
|
|
4G0F
 
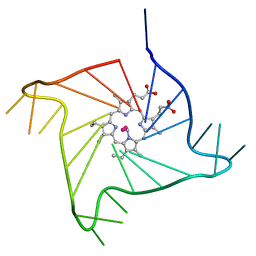 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P6) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
4FXM
 
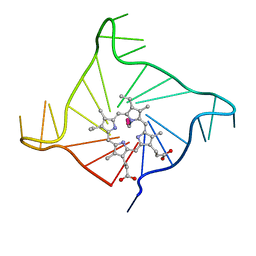 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P21212) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-03 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
3QP4
 
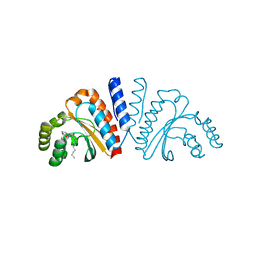 | | Crystal structure of CviR ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
