4DAG
 
 | |
3MAW
 
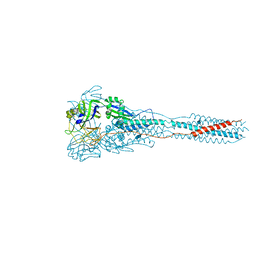 | |
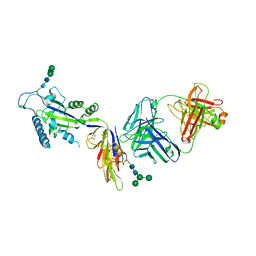
1SEB
 
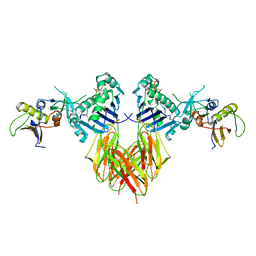 | | COMPLEX OF THE HUMAN MHC CLASS II GLYCOPROTEIN HLA-DR1 AND THE BACTERIAL SUPERANTIGEN SEB | | Descriptor: | ENDOGENOUS PEPTIDE MODEL, POLY-ALA, ENTEROTOXIN TYPE B, ... | | Authors: | Jardetzky, T.S, Brown, J.H, Gorga, J.C, Stern, L.J, Urban, R.G, Chi, Y.I, Stauffacher, C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1995-11-26 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen.
Nature, 368, 1994
|
|
5T1D
 
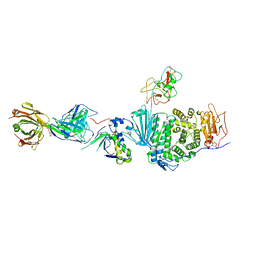 | | Crystal structure of EBV gHgL/gp42/E1D1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E1D1 IgG2a heavy chain, E1D1 IgG2a light chain, ... | | Authors: | Sathiyamoorthy, K, Jardetzky, T.S. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins.
Nat Commun, 7, 2016
|
|
8TQI
 
 | |
8TQK
 
 | |
4WSG
 
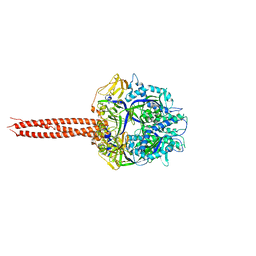 | | Crystal Structure of Soluble WR PIV5 F-GCNt | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Poor, T.A, Song, A.S, Welch, B.D, Kors, C.A, Jardetzky, T.S, Lamb, R.A. | | Deposit date: | 2014-10-27 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Soluble WR PIV5 F-GCNt
J.Virol., 89, 2015
|
|
4JF7
 
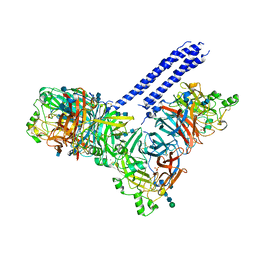 | | Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Welch, B.D, Yuan, P, Bose, S, Kors, C.A, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5018 Å) | | Cite: | Structure of the Parainfluenza Virus 5 (PIV5) Hemagglutinin-Neuraminidase (HN) Ectodomain.
Plos Pathog., 9, 2013
|
|
5EJB
 
 | | Crystal structure of prefusion Hendra virus F protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Wong, J.W, Jardetzky, T.S, Paterson, R.G, Lamb, R.A. | | Deposit date: | 2015-11-01 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and stabilization of the Hendra virus F glycoprotein in its prefusion form.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2P6A
 
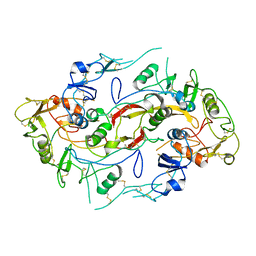 | | The structure of the Activin:Follistatin 315 complex | | Descriptor: | Follistatin, Inhibin beta A chain, probable fragment of follistatin | | Authors: | Lerch, T.F, Shimasaki, S, Woodruff, T.K, Jardetzky, T.S. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biophysical coupling of heparin and activin binding to follistatin isoform functions.
J.Biol.Chem., 282, 2007
|
|
3QW9
 
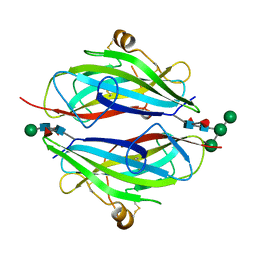 | | Crystal structure of betaglycan ZP-C domain | | Descriptor: | Transforming growth factor beta receptor type 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lin, S.J, Jardetzky, T.S. | | Deposit date: | 2011-02-27 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-{beta} binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PHF
 
 | |
6UQR
 
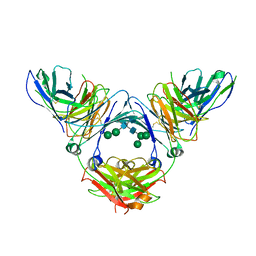 | | Complex of IgE and Ligelizumab | | Descriptor: | IgE, Ligelizumab, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tarchevskaya, S.S, Kleinboelting, S, Jardetzky, T.S. | | Deposit date: | 2019-10-21 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6502912 Å) | | Cite: | The mechanistic and functional profile of the therapeutic anti-IgE antibody ligelizumab differs from omalizumab.
Nat Commun, 11, 2020
|
|
3TSI
 
 | | Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) stalk domain | | Descriptor: | Hemagglutinin-neuraminidase | | Authors: | Bose, S, Welch, B.D, Kors, C.A, Yuan, P, Jardetzky, T.S, Lamb, R.A. | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion.
J.Virol., 85, 2011
|
|
3T1E
 
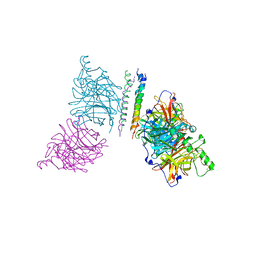 | | The structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a 4-helix bundle stalk | | Descriptor: | Hemagglutinin-neuraminidase | | Authors: | Yuan, P, Swanson, K, Leser, G.P, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-07 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
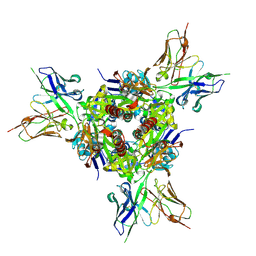
5U68
 
 | |
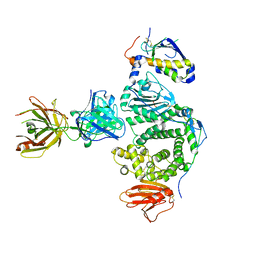
5W0K
 
 | |
8EAY
 
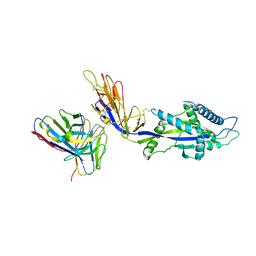 | | HMPV F complex with 4I3 Fab | | Descriptor: | 4I3 heavy chain protein, 4I3 light chain protein, Fusion glycoprotein F0 | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
8E2U
 
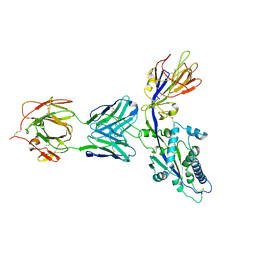 | | HMPV F monomer bound to RSV-199 Fab | | Descriptor: | Fusion glycoprotein F0, RSV-199 heavy chain, RSV-199 light chain | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
8EBP
 
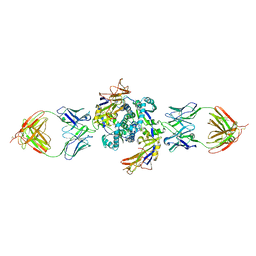 | | HMPV F dimer bound to RSV-199 Fab | | Descriptor: | Fusion glycoprotein F0, RSV-199 heavy chain protein, RSV-199 light chain protein | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
8DZW
 
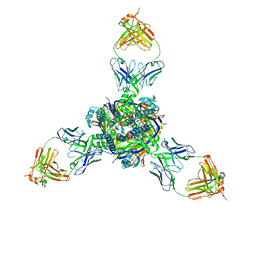 | | RSV F trimer bound to RSV-199 Fab | | Descriptor: | RSV fusion protein, RSV-199 Heavy chain protein, RSV-199 Light chain protein | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
5HYS
 
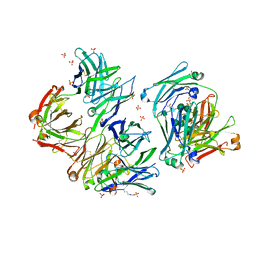 | | Structure of IgE complexed with omalizumab | | Descriptor: | Epididymis luminal protein 214, Ig epsilon chain C region, SULFATE ION, ... | | Authors: | Pennington, L.F, Tarchevskaya, S.S, Sathiyamoorthy, K, Jardetzky, T.S. | | Deposit date: | 2016-02-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of omalizumab therapy and omalizumab-mediated IgE exchange.
Nat Commun, 7, 2016
|
|
4FZH
 
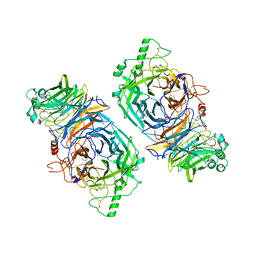 | | Structure of the Ulster Strain Newcastle Disease Virus Hemagglutinin-Neuraminidase Reveals Auto-Inhibitory Interactions Associated with Low Virulence | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-neuraminidase | | Authors: | Yuan, P, Paterson, R.G, Leser, G.P, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5008 Å) | | Cite: | Structure of the ulster strain newcastle disease virus hemagglutinin-neuraminidase reveals auto-inhibitory interactions associated with low virulence.
Plos Pathog., 8, 2012
|
|
4GRG
 
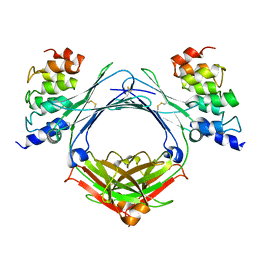 | |
4GT7
 
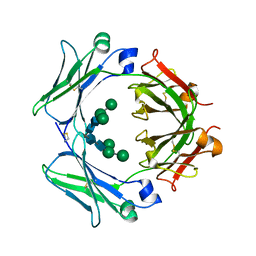 | | An engineered disulfide bond reversibly traps the IgE-Fc3-4 in a closed, non-receptor binding conformation | | Descriptor: | Ig epsilon chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wurzburg, B.A, Kim, B.K, Jardetzky, T.S. | | Deposit date: | 2012-08-28 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An engineered disulfide bond reversibly traps the IgE-Fc3-4 in a closed, nonreceptor binding conformation.
J.Biol.Chem., 287, 2012
|
|
